Introduction and setup
Overview
Teaching: 15 min
Exercises: 15 minQuestions
What will we be learning?
How will we be learning?
Objectives
Review schedule
Set expectations for engagement and behavior
Introduce ourselves
Acknowledgement of country
We wish to acknowledge the custodians of the land we reside on, for the developers of these lessons, these are the Wadjuk (Perth region) people of the Nyoongar nation. We would like to pay our respect to their Elders past, present and emerging and we acknowledge and respect their continuing culture and the contribution they make to the life of this city and this region.
Schedule
| AEST | AWST | Event | Notes |
|---|---|---|---|
| 09:30 | 07:30 | Introduction and setup | |
| 10:00 | 08:00 | Scientific Computing | |
| 11:00 | 09:00 | break | |
| 11:30 | 09:30 | Best Practices In Computing | |
| 12:30 | 10:30 | break | |
| 13:30 | 11:30 | Data Management | |
| 14:30 | 12:30 | break | |
| 15:00 | 13:00 | Project Management | |
| 16:00 | 14:00 | end |
Content
This workshop is designed to give you training in a broad range of skills that will be useful for your career in academia and beyond. We are not aiming to provide a deep dive into any of the material covered here, but instead will focus on making sure that everyone comes away with enough information to know what skills are useful, what questions to ask, and where to go for more information.
If you are interested in further engagement please see the links within the lessons, the bonus content lesson (not part of the HWSA workshop), chat to us in the breaks, or reach out via email or slack to get some more advice from the ADACS team.
Engagement
This workshop is developed for three main delivery methods:
- Facilitated, in person, at the HWSA venue in Tasmania
- Facilitated, online, in parallel with (1)
- Self-paced, online, via this website.
For the HWSA school we will set up a shared google doc for people to contribute to as we go along. This means that contributions can be anonymous and asynchronous as needed. Whilst we have ADACS people here to facilitate the workshop, we encourage people to ask for help from their peers, and to offer help to those that may need it.
This workshop is all about learning by doing. We will be engaging in live coding type exercises for most of the workshop, and we will set challenges and exercises for you to complete in groups. The more you engage with your fellow learners and the more questions that you ask, the more that you will get out of this workshop.
We will be using sticky notes for in-person participants to indicate their readiness to move on: please stick them on your laptop screen to indicate if you need help or are done and ready to move ahead. For those joining online we’ll be using the cross or check icons (zoom) to indicate the same.
We will use a shared document (etherpad) to manage and record many of our interactions.
Conduct
This workshop will be an inclusive and equitable space, which respects:
- the lived experience of it’s attendees,
- the right for all to learn, and
- the fact that learning means making mistakes.
We ask that you follow these guidelines:
- Behave professionally. Harassment and sexist, racist, or exclusionary comments or jokes are not appropriate.
- All communication should be appropriate for a professional audience including people of many different backgrounds.
- Sexual or sexist language and imagery is not appropriate.
- Be considerate and respectful to others.
- Do not insult or put down other attendees.
- Critique ideas rather than individuals.
- Do not engage in tech-shaming.
See the ASA2022 code of conduct and the Software Carpentries code of conduct for more information.
Introduce yourselves
Introduce yourself to your peers by telling us your name, a game you enjoy playing, and a food that you would like to try but never have.
Please also introduce yourself in more detail in the shared google doc.
Key Points
This is not a lecture
The more you engage the more you will benefit
Everyone is here to learn and that means making mistakes
Scientific Computing
Overview
Teaching: 30 min
Exercises: 30 minQuestions
What is scientific computing?
How can I automate common tasks?
What is reproducible research?
Objectives
Feel good about your computing goals
Appreciate various levels of automation
Understand what reproducibility means
Scientific Computing
Scientific computing is best understood by its relation to, and difference from, computer science, and software engineering. Computer science places a big focus on correctness of the code and the efficiency of the algorithm. Software engineering places a big focus on the usability of the software, and how it fits within an ecosystem of other solutions. Scientific computing cares little for any of these concerns, and is primarily concerned with producing correct scientific results. There is of course a large overlap between the three ideals as shown in the ternary diagrams below.
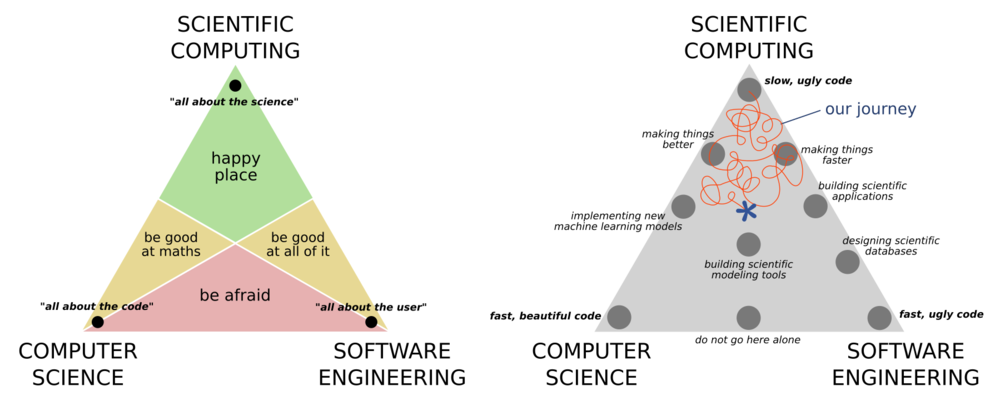
It is rare today that you can work on any project without having to some kind of computing. Many physics and astronomy students are not required to take any computing courses as part of their undergrad (though many do). Thankfully this is changing, however the courses are often focused on software engineering or computer science rather that scientific computing.
Your computing journey
In small groups, briefly share your computing journey, and where you feel that you currently sit in the above diagram.
As you discuss make notes in our shared document along the following topics:
- When writing code or developing software, what is your primary concern?
- What formal training have you had?
- What are some good resources for self/peer learning?
- What languages / tools do you commonly use or find particularly useful?
A key takeaway from the above is that you should not hold yourself to the standards of a computer scientist or a software engineer because you are neither of these things. Your goal is not to make software that is widely usable, highly performant, or scalable. First and foremost you are focusing on getting the science correct, and the rest is a bonus. Having said this, there is of course a large overlap between the three disciplines and some important lessons that we can take from software engineers and computer scientists that will make our science results more robust, more easy to replicate, easier to adapt and reuse, as well as allowing us to take on larger projects with more ambitious goals. This will be the focus of our first two lessons today.
Automate your way to success
Most of the activity in which we engage on a daily basis in research is computer based. A typical researcher will use a combination of graphical and command line user interfaces (GUIs and CLIs) to interact with this software. Each have their tradeoffs and strengths, but a major difference is that it is often easy to automate programs that have a command line interface, whilst it is difficult to do the same with a graphical interface.
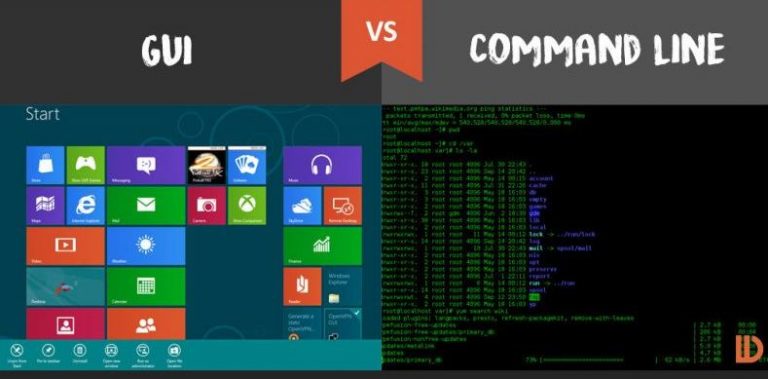
Your day-to-day sciencing workflow
Think of the tasks that you do as part of your daily science-ing, and the tools that you use to make this happen. Discuss the following with your colleagues and then write in the shared document.
- Match your activity to a tool that you use, or one that you want to use.
- For each of the tools that you have identified note if it has a GUI or CLI (or both).
- What aspects of your day-to-day do you wish you could automate?
In our shared document contribute one item from each of the above.
Simple tasks are often the most easily automated. For example, maybe you want to back up the data on your (linux) computer and you have purchased an additional hard drive for this very purpose. Due to previous bad experience with RAIDed hard drives, an aversion to over complicated software solutions, and no interest in offsite or incremental backups, you have decided that your backup and recovery will consist of copying files from one hard drive to another on a regular basis. For this task you put a reminder in your calendar every monday to execute the following code:
rsync -av --delete /data/working/${USER} /data/backup/${USER}
This will use the rsync program to copy data from the hard drive mounted at /data/working/${USER} to the one mounted at /data/backup/${USER}.
Here ${USER} is the bash variable which refers to your username.
The options -av will preserve the file ownership, permissions, and access date/time, and --delete will make sure that files that are in the working directory but not in the backup directory will be deleted.
Essentially this will create an exact mirror of one drive onto another.
The program rsync is smart enough to do this mirroring by only copying the changes from one drive to another so that you don’t have to delete and re-copy the data every week.
Challenge
- What are some of the draw backs in using this method of backup?
- How could you ensure that you use the same command each time you do your backup?
- How could you automate this task so that you no longer have to think about it or spend time on it?
Solution notes
- If you don’t have your calendar open you might forget to run the program.
- You might not run the program on every Monday because you don’t work every Monday.
- Keep the above text in a file called
backup_instructions.txtso that you remember how to do it correctly.- Convert
backup_instructions.txtintodo_backup.sh, add a#!(shebang) line, and make the file executable, so you can type./do_backup.sh- You could automate by adding the above line to your crontab so that it will always run on a Monday morning without your input
Any work that you can do from the command line can be automated in a similar way. A good approach to take when considering automating a task is:
- Complete the task manually, using the command line
- Copy all the commands that you used into a file so that you have an exact record
- Save the file as a
.sh(OSX/Linux) or.batfile (Windows) - You now have an automation script!
- Next time you need to do said task, just run the script
Optional extras to make your automation even better include:
- Adding some command line arguments to your script so that you can do slightly different things each time
- Generalize the script so that information that may need to change between runs can be automatically generated or calculated
The Linux ecosystem is built on the idea of having a large number of tools each with a very specific purpose. Each tool should do one thing and do it well. There are many seemingly simple tools that already exist on your Linux (and OSX, or WSL) computer that you can chain together to great effect.
Favorite tools
- Discuss with your group some of the command line tools that you use most often.
- Let people know what tool you wish existed (because it might already!).
- Share your thoughts in our shared document.
- Have a look at the with list of others and see if you can propose a solution or partial solution.
Let’s explore some of these useful command line tools that you might use to provide some low-level automation to your life.
grep
wget
curl
sed
cut
wc
bc
We’ll explore these together, but if at a later point you want more help on these tools you can:
- use program help via the
--helpflag - use manual pages like this
man grep,man cut, or evenman man - ask dr google
Reproducible research
Reproducibility is the idea of being able to re-run your analysis with the same data and get the same results. Related concepts are replicability, robustness, and generalisabiltiy, as demonstrated in the following diagram.
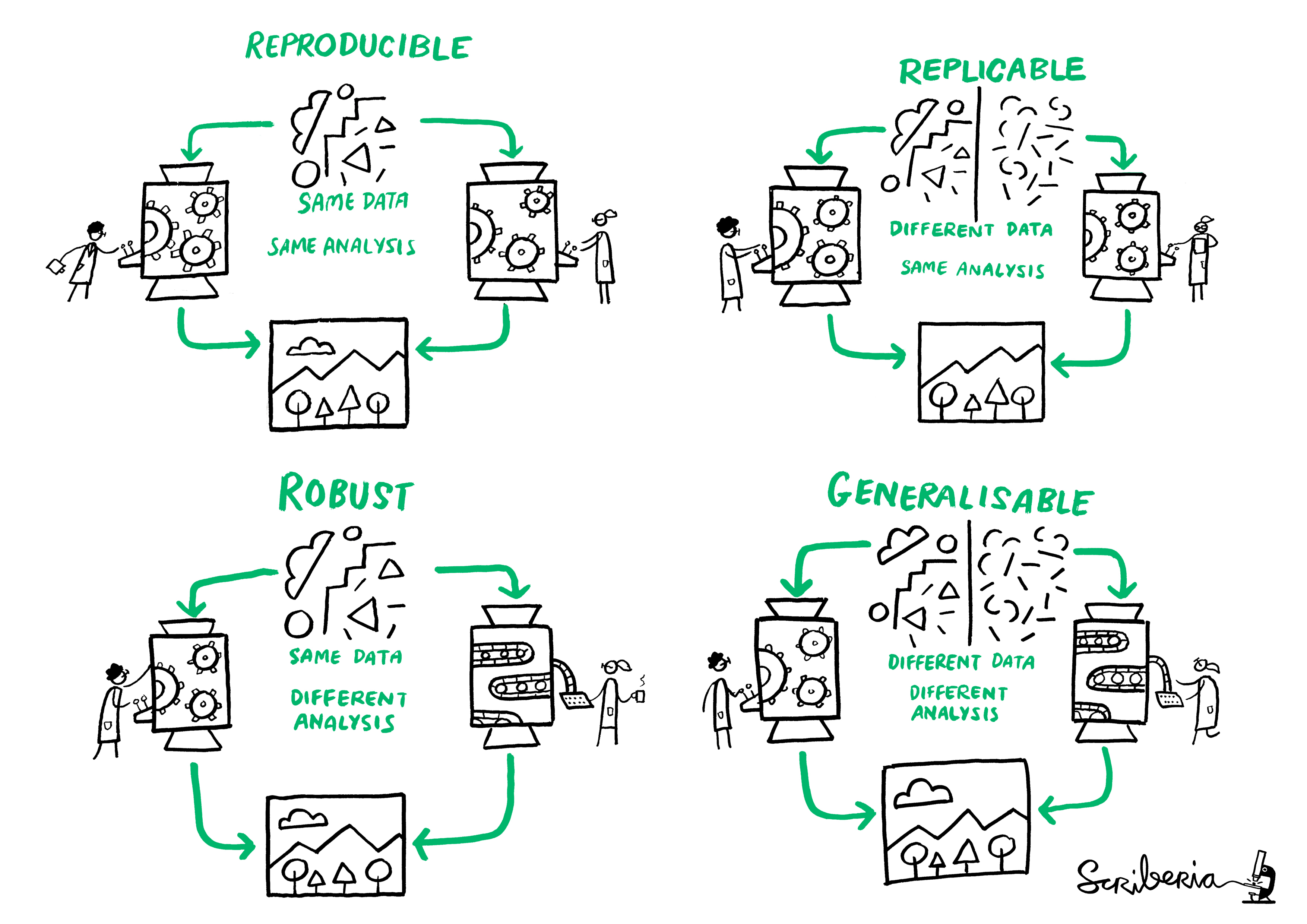
There are many reasons why you would re-run your analysis or data processing:
- As you develop your data collection, processing, and analysis techniques you will make changes to the process. Each change means re-doing all the work again.
- Your work may be extended or expanded as new data become available, requiring you to apply the same processing to different data,
- During the publication review process you need to make adjustments to the data selection or processing based on collaborator or referee’s comments,
- Other people may want to apply your techniques to their data and compare results.
A science workflow that can be consistently re-run and easily adapted is of huge benefit to you, the researcher. Sharing your data, and your methodology (via code and documentation) with others can also benefit the community because it will allow your work to be rerun and adapted, saving others time whilst generating citations and accolades for you.
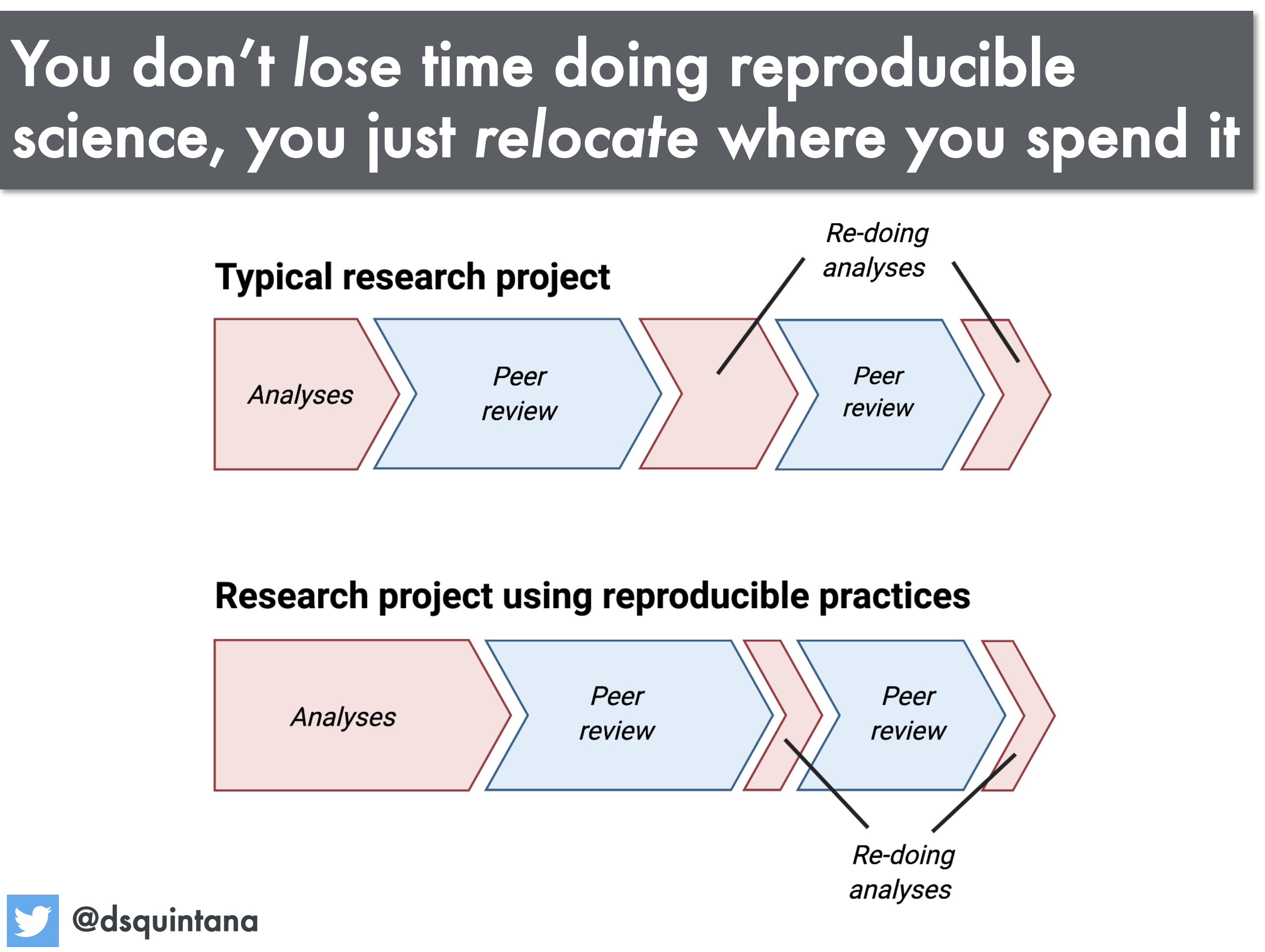
Discussion
- If you were to redo all your data processing from scratch, how confident are you that you would get the same result?
- What aspects of your workflow have you automated?
- What parts of your workflow cannot be automated, and what can you do about that?
- Is automation essential for reproducible research?
As always, share your thoughts with your group and then put some notes into the shared document.
Key Points
Scientific computing has different goals from computer science and software engineering
You are a scientist, your focus is on the science
Automation can occur at many levels
Reproducibility is good for everyone (especially you)
Morning Tea
Overview
Teaching: min
Exercises: minQuestions
Objectives
Time for our first break.
Key Points
Best Practices In Scientific Computing
Overview
Teaching: 30 min
Exercises: 30 minQuestions
What does best practice mean?
What should I be doing?
What should I try to avoid?
Objectives
Appreciate guidelines for best practice
Work through an exercise demonstrating some of these practices
Be less scared about testing and documentation
Best Practices In Scientific Computing
While computer science and software engineering have different goals than scientific computing, many of the best practices in these disciplines are still applicable to scientific computing.
Since the top priority of scientific computing is to have software that produces correct results, we can make our lives easier by adopting practices that make our code or scripts easier to understand (by humans) so that errors can be found and fixed easily. Additionally, since our research work is continuously changing it is very likely that we will revisit our scripts to re-use them (in part or in whole) or expand their use. Here again readability and clarity will be of benefit, but so will version control, and modularization.
Here are some guiding principles that should be followed when planning or writing scripts, regardless of language:
| Guideline | Description |
|---|---|
| Clarity over efficiency | Prioritize writing clear code over efficient code. Make use of programming idioms1. Do standard things in standard ways. Write code for humans to understand in the first instance, and only secondarily for computers. |
| Naming is important | Choose meaningful and useful names for variables/functions/classes/scripts. Typically: objects are nouns, functions are verbs, pluralize variables that represent lists or collections. Unhelpful names can be annoying to work with, whilst confusing names can be downright destructive. |
| Don’t repeat yourself | When something works reuse it. Bundle repeated code into functions and call those functions. Bundle functions into modules that can be imported. Write simple (single intent) tools that can be easily incorporated into larger workflows. |
| Don’t repeat others | If something seems routine then there is almost always an existing solution that you can rely on. Look for existing libraries/modules or programs that you can make use of. Don’t reinvent the wheel2 |
| Document | You will forget what you did and why you did it. Write yourself a document that describes the problem you were trying to solve, the approach that you took, and how you solved it. If the solution is a script, then describe how to use the script including the inputs, what options are available, and what the output is. This can be a README.md file, docstrings (in python) or a pdf. The format is less important than the fact that the documentation exists. |
| Test | Only the very lucky get things right the first time. Don’t rely on luck. When you write a script, do something to convince yourself that it works. Manually inspecting results for a known example is form of testing. This of testing as validation. |
| Version control | When moving towards a solution we often make a wrong turn. Use a version control system to create a ‘checkpoint’ or ‘save point’ that you can easily come back to if things go bad. You don’t need to do pull requests, branching, merging, or upload your files to GitHub for version control to be useful. |
| Avoid premature optimization | Optimization can save time in the term run but always costs time in the short term. Optimize your time by firstly solving the problem, and only engage in optimization after you find out that your code is taking too long or using too many resources3. |
In this lesson we will focus on repetition, version control, testing, documentation, and repetition. To demonstrate the utility of these topics we’ll be working on a common task - analyzing meteorite falls around the world.
Use case - visualizing a data set
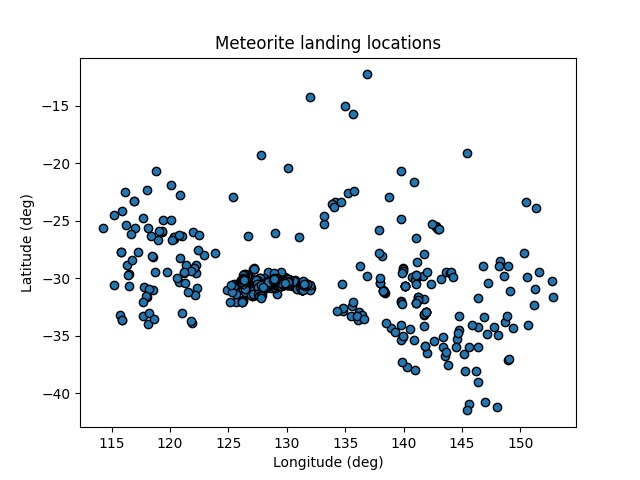
Let’s go through the process of visualizing some data and see if we can incorporate the above best practices as we go. We will be working with a data file that contains information about meteorites that have fallen in Australia, and our aim is to plot the location of the meteorites, and give an indication of their masses.
To begin with we need to obtain the data. We have a nice data set prepared for you which you can download the data with the command4:
wget https://raw.githubusercontent.com/ADACS-Australia/HWSA-2022/gh-pages/data/Australian_Meteorite_Landings.csv
# or if the above didn't work, then try
curl -O https://raw.githubusercontent.com/ADACS-Australia/HWSA-2022/gh-pages/data/Australian_Meteorite_Landings.csv
We will start with a peak inside this file with the head command:
head Australian_Meteorite_Landings.csv
output
id,mass (g),reclat,reclong 5051,488.1,-33.15639,115.67639 48653,324.0,-31.35,129.19 7743,30.0,-31.66667,152.83333 10033,127.0,-29.46667,151.61667 10120,26000.0,-33.35,146.85833 12264,41730.0,-35.08333,139.91667 16643,330000.0,-26.45,120.36667 16738,8887.5,-40.975,145.6 16766,11300.0,-29.8,141.7
From this you can see that we have given you a .csv file recording meteorites that have landed in Australia.
The four columns represent an ID, a mass, and a recorded position in latitude and longitude.
We’ll use python for our visualization, so we begin by reading the file with python. Reproducing the above quick summary in python we would write a script like the following:
# the csv library has a lot of read/write functions for csv files
import csv
csv_list = []
# opening the CSV file
with open('Australian_Meteorite_Landings.csv', mode ='r') as file:
# read the CSV file
csv_reader = csv.reader(file)
# loop over the lines in the file and add each to our list
for line in csv_reader:
csv_list.append(line)
# print the first 10 lines to ensure that we are on the right track.
print(csv_list[:10])
output
[['id', 'mass (g)', 'reclat', 'reclong'], ['5051', '488.1', '-33.15639', '115.67639'], ['48653', > '324.0', '-31.35', '129.19'], ['7743', '30.0', '-31.66667', '152.83333'], ['10033', '127.0', '-29.> 46667', '151.61667'], ['10120', '26000.0', '-33.35', '146.85833'], ['12264', '41730.0', '-35.08333', > '139.91667'], ['16643', '330000.0', '-26.45', '120.36667'], ['16738', '8887.5', '-40.975', '145.6'], > ['16766', '11300.0', '-29.8', '141.7']]
At this point we should save our work in a file and make sure that we are on the right track by running the script and inspecting the output. Since we have just started our new project and have some initial progress, this is a great time to set up our version control.
Start a new project
- Create a new directory for your project with a name that makes sense to you
- Save your initial python script as
plot_meteorites.py(what we’ll eventually be doing)- Run your script and ensure that it’s not broken
- Initialize a git repository in this directory by typing
git init- Tell git that you want this new file to be tracked by using
git add <filename>- Also commit your data file using the same technique
- Save your initial progress by creating a new commit to your repository via
git commit -m > <message>
- The first commit message can be something simple like “initial version”
- Check that you have committed your progress by running
git log
If at any point we are editing our work and we break something, or change our mind about what we are doing, so long as we have the files under version control we can go back to our previous save point using:
git checkout -- <filename>
If we want to reach way back in time we can do
git checkout <hash> <filename>
Where the <hash> is one of the long alphanumeric strings that are shown when we run git log.
Having good commit messages will make it easier to tell which commit we should be going back to.
Now we have some first step that we can come back to later if we mess things up. Let’s try and do something with this data - computing the mean mass of the meteorites. We can do this by adding the following to our script and re-running:
import numpy as np
all_mass = []
# csv_list[1:] will skip the first line which is our header (not data)
for id, mass, lat, long in csv_list[1:]:
# skip missing data
if mass != "":
# Convert string -> float and append to our list
all_mass.append(float(mass))
# Output mean of the masses
print("The mean mass is:", np.mean(all_mass), "g")
...
The mean mass is: 80919.38811616955 g
Save your progress
- Once our script is in a working state, save it, add it to git, and then commit it
- Use a commit message that describes what we did in less than 50 chars
- Double check your git log to see that the changes have been applied
The above is fine but that was a bit of work for what feels like a very standard task. Let’s be guided by the “don’t repeat others” mentality, and see if we can find an existing solution that will do this work for us. The python data analysis library pandas has a lot of great functionality built around data structures called data frames which are a fancy kind of table. So if we let pandas do all the hard work for us then we can extend our above code with the following:
# Use the pandas library
import pandas as pd
# read a csv file into a data frame
df = pd.read_csv('Australian_Meteorite_Landings.csv')
# Have a look at the data
print("csv data\n--------------------------")
print(df)
# Use pandas to do some quick analysis
print("\npanda describe\n--------------------------")
print(df.describe())
output
... csv data -------------------------- id mass (g) reclat reclong 0 5051 488.1 -33.15639 115.67639 1 48653 324.0 -31.35000 129.19000 2 7743 30.0 -31.66667 152.83333 3 10033 127.0 -29.46667 151.61667 4 10120 26000.0 -33.35000 146.85833 .. ... ... ... ... 638 30359 262.5 -32.03333 126.17500 639 30361 132000.0 -14.25000 132.01667 640 30362 40000.0 -31.19167 121.53333 641 30373 118400.0 -29.50000 118.75000 642 30374 3800000.0 -32.10000 117.71667 [643 rows x 4 columns] panda describe -------------------------- id mass (g) reclat reclong count 643.000000 6.370000e+02 643.000000 643.000000 mean 19219.513219 8.091939e+04 -30.275400 130.594478 std 14869.941958 1.029442e+06 2.940091 7.655877 min 471.000000 0.000000e+00 -41.500000 114.216670 25% 10136.500000 3.280000e+01 -30.841665 126.587915 50% 15481.000000 1.291000e+02 -30.383330 128.916670 75% 23537.500000 2.426000e+03 -30.086415 132.008335 max 56644.000000 2.400000e+07 -12.263330 152.833330
With only a few lines we can load the data and have a quick look. You can see that the count of mass is only 637 out of 643 so pandas has recognized that there is missing mass data and has even calculated a mean mass for us.
Save your progress
- Once our script is in a working state, save it, add it to git, and then commit it
- Use a commit message that describes what we did in less than 50 chars
- Double check your git log to see that the changes have been applied
Now that pandas is doing all the file reading and format handling for us we can do away with our previous attempt at reading a csv file.
Delete some code and check it in
- Delete all the code before the “import pandas as pd” command
- Run
git statusto see which files have changed- Run
git diff plot_meteorites.pyto see what changes have been made to the file- Run the script and confirm that it still works
- Add and commit our work to our git repository
Pre-existing solutions
See if you can identify a python package (module) that will help you with each of the following tasks:
- Read
.fitsformat images- Create a ‘corner plot’ from multi-dimensional data
- Plot images using sky coordinates using the correct projection
- Read and write
.hdf5format data files- Solve equations analytically/exactly
- Train and evaluate machine learning models
Head over to the python package index at pypi.org, and search for the packages you found.
In our shared document make a note of a package that you wish existed, and then look at the wish list of others and see if you can suggest a (partial) solution.
In the above challenge you may have seen a few modules that occur frequently, either because they are large and multi-purpse modules, or because they are fundamental and used as a building block for many others.
For example, scipy and numpy are fundamental to most of the scientific computing libraries that are built, and packages like astropy are becoming very widely use in astronomy.
If you have noticed a bias towards python in our teaching examples, it is because of this rich ecosystem of freely available and easy to use modules. The large adoption within the astronomy community, also means that it is easy to get relevant and specific help from your peers. As you work in other fields you’ll notice that different software packages are standard and usually for the same reason - easy of access, and access to support. At the end of the day the right tool is the one that gets the job done, and you should not be afraid of exploring beyond python to get your work done.
Visualizing data
Back to our main task of making a nice plot of our data. Let’s make a scatter graph of the locations of the meteorites using the matplotlib module.
Since this should be a simple task, we shouldn’t be supprized to find that we can complete it in a few lines of code. Add the following to our script and run it:
# import the plotting library with a short name
import matplotlib.pyplot as plt
# make a scatter plot
plt.scatter(df['reclong'], df['reclat'], edgecolors='0')
# label our plot!
plt.xlabel("Longitude (deg)")
plt.ylabel("Latitude (deg)")
plt.title("Meteorite landing locations")
# pop up a window with our plot
plt.show()
Our first plot
Save your progress
- Commit your changes to the plotting script.
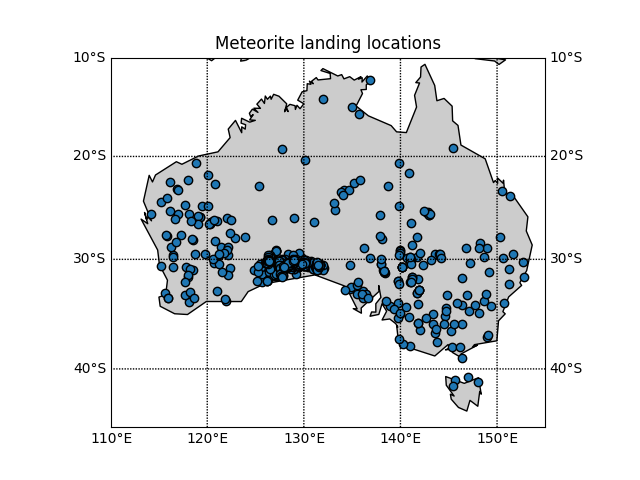
Our plot looks vaguely Australia shaped but to be sure lets plot this against a map of Australia to make sure we believe the locations. Doing this manually would be difficult so, after some Googling, we have found a python module that can do it for you called Basemap
Replace the previous plotting section of your script with the following:
from mpl_toolkits.basemap import Basemap
import numpy as np
import matplotlib.pyplot as plt
fig=plt.figure()
# setup map projection
bm = Basemap(
llcrnrlon=df['reclong'].min()-5, # Lower left corner longitude
urcrnrlon=df['reclong'].max()+5, # Upper right corner longitude
llcrnrlat=df['reclat'].min()-5, # Lower left corner latitude
urcrnrlat=df['reclat'].max()+5, # Upper right corner latitude
)
# Draw in coasline and map lines
bm.drawcoastlines()
bm.fillcontinents()
bm.drawparallels(np.arange(-90,90,10), # The location of the parallels
labels=[1,1,0,1]) # Which axes to show labels on
bm.drawmeridians(np.arange(-180,180,10),labels=[1,1,0,1])
# Convert your data to the Basemap coordinates and add it to the plot
x, y = bm(df['reclong'], df['reclat'])
plt.scatter(x, y,
edgecolors='0',
zorder=2) # make the points be in front of the basemap
plt.show()
Our second plot
Save your progress
- Commit your changes to the plotting script.
Creating a reusable script
This is looking great but before we go any further we should make the plotting component into a function as part of a script so it’s easier to rerun for different locations on the Earth. Moving commonly used code into a function, rather than just copy/pasting it, is ane example of don’t repeat yourself.
Let’s take all of the plotting part of our script and put it into a function called plot_meteor_locations, and allow the user to supply a data frame with the data, and a bounding box of lat/long for the plotting area.
Since this function is something that we’ll be using often, it’s a good idea to write some documentation for the function.
In python we can use a docstring to show what the function is supposed to do and what the expected inputs ared.
Alternatively we could write a short noted in a REDEME.md file that describes how to use the program.
import matplotlib.pyplot as plt
from mpl_toolkits.basemap import Basemap
import pandas as pd
import numpy as np
def plot_meteor_locations(df):
fig=plt.figure()
# setup map projection
bm = Basemap(
llcrnrlon=df['reclong'].min()-5, # Lower left corner longitude
urcrnrlon=df['reclong'].max()+5, # Upper right corner longitude
llcrnrlat=df['reclat'].min()-5, # Lower left corner latitude
urcrnrlat=df['reclat'].max()+5, # Upper right corner latitude
)
# Draw in coasline and map lines
bm.drawcoastlines()
bm.fillcontinents()
bm.drawparallels(np.arange(-90,90,10), # The location of the parallels
labels=[1,1,0,1]) # Which axes to show labels on
bm.drawmeridians(np.arange(-180,180,10),labels=[1,1,0,1])
# Convert your data to the Basemap coordinates and add it to the plot
x, y = bm(df['reclong'], df['reclat'])
plt.scatter(x, y,
edgecolors='0',
zorder=2) # make the points be in front of the basemap
plt.show()
return
if __name__ == '__main__':
# read a csv file into a data frame
df = pd.read_csv('Australian_Meteorite_Landings.csv')
plot_meteor_locations(df)
Now you have a script that is easy to rerun if you want to recreate the plot and if anyone asks you how you generated the results you can show them your script so you can go through the steps one by one to confirm their validity.
Let’s focus on New Zealand
- Verify that your code works and then commit the changes to your repo
- Download the New Zealand data file
- Edit your script to use the new data file, save, and rerun
Making your script re-usable
Right now if someone wants to use your script they would have to import that function into their own script like this:
from plot_meteorites import plot_meteor_locations
# ... load a data frame called df
plot_meteor_locations(df)
However, it is often nice to have a basic command line interface.
In python any arguments that we pass to the program are stored in a list, and we can access this list from the sys module via sys.argv.
If we want to read the name of the data file from the command line we can modify our if __main__ clause like this:
import sys
# ... the rest of our code
if __name__ == '__main__':
# get the filename from the command line as the last argument
csv_file = sys.argv[-1]
# read a csv file into a data frame
df = pd.read_csv(csv_file)
plot_meteor_locations(df)
Here the sys.argv[-1] refers to the last argument that was given.
It will always be interpreted as a string, and in this case we’ll use it as a filename.
add a command line interface
- modify your code so that it reads the name of the data file from the command line
- test your code by running it on both the Australia and New Zealand data
- commit your changes to your repository (including the new data file)
Testing
Testing is something that many people think is too much work to bother with. However, we are always testing our code even if we don’t think of it that way. Any time you run your code and compare the behavior/output with expectations is a test. Testing is not hard, and we already do it so let’s just embrace that.
In the previous challenge we ran our code on two different data sets, and then inspected the output to confirm that the code was working as intended. To do that we ran the following bash commands:
python3 plot_meteorites.py New_Zealand_Meteorite_Landings.csv
python3 plot_meteorites.py Australian_Meteorite_Landings.csv
If we were to make some changes to our code, it is a good idea to re-run these tests to make sure that we haven’t broken any of the existing functionality.
If we are adding some new functionality, or fixing some bug, then it would be good to have a new test to run to verify that the new parts of the code are working as intended.
To remind ourselves of how we do the testing lets make a file to record the process.
In fact, we can just make a file called test.txt which includes the above two lines of code and maybe a short description of what is expected.
When we want to run the test we just open the file and follow the instructions, copy/paste the lines of code as required.
If we think back to our lesson on scientific computing and automation we can take this a step further.
We could write test.sh with the same lines of code as test.txt (and convert the instructions into comments), and then use source test.sh to have all the code execute automatically.
As your testing becomes more involved you’ll eventually want to look into some more automated ways of not just running the tests, but ensuring that the tests all pass.
For this you’ll have to come up with some quantitative measures of what pass/fail look like, and then write a bit of code that will check against these conditions.
If/when you get to this point you should start thinking about using a testing framework such as pytest to help with the automation and organization of the tests.
(You still have to write them yourself though!)
Create test file
- Create either
test.txtortest.shusing the example above- Do/run the test
- Commit the test file to your git repo
Documentation
Documenting your code is important as it makes it clear how you code generates your results and shows others (and yourself) how to use your code. There are several levels of documentation and what level is appropriate depends on the purpose, complexity and who will use your code.
Documentation levels include:
- Code comments
- README.md
--helpor docstrings (Python)- readthedocs.io which can host your documentation and using sphinx you can automate it’s generation
- A user guide (pdf or markdown) to explain how to use the code with explanations and common examples
Considering your audience is a good indicator of what documentation level you should aim for
- You in 6 months time (level 1-3)
- Other developers of the code (level 1-4)
- Other users (level 4-5)
Markdown (.md) is a common syntax to generate documentation, for example a README.md will automatically be rendered on GitHub.
Lets create a README.md for your code.
# Meteorite Analysis Software
This git repository contains python scripts to analyze meteorite data for my research, which is bound to win me the noble prize.
## Installation
This software isn't installable (yet), but it's always good to describe how to install your software. Some common ways to install python scripts include
`python setup install`
or
`pip install .`
## Data
It is always best to note where your data came from and describe how to use it.
### Australian_Meteorite_Landings.csv
Data downloaded from https://raw.githubusercontent.com/ADACS-Australia/HWSA-2022/gh-pages/data/Australian_Meteorite_Landings.csv.
A truncated version of the data from https://data.nasa.gov/Space-Science/Meteorite-Landings/ak9y-cwf9.
id: NASA meteorite
mass (g): Mass of the meteorite in grams
reclat: Recorded latitude in degrees
reclong: Recorded longitude in degrees
## Running Software
Create analysis plots using:
`python plot_meteorites.py`
You can use VSCode extensions to view the rendered version or push it to GitHub to view it on the webpage. This will remind you how to use your code and where you got your data.
Create a README file for your repo
- Create a
README.mdfile in your repo- Add your name, an description of the intent of the repo, and what the various files do
- Commit this file to your git repo
-
See programming idioms ↩
-
Re-inventing the wheel can be a great learning experience, however when you are focusing on getting work done, it’s most often not a good use of your time. ↩
-
wgetisn’t available via gitbash, butcurlis so windows users may have to usecurl↩
Key Points
Validation is testing
Documentation benefits everyone (especially you)
Version control will save you time and effort
Lunch
Overview
Teaching: min
Exercises: minQuestions
Objectives
Time for Lunch.
Key Points
Managing Data
Overview
Teaching: 30 min
Exercises: 30 minQuestions
Why do I need to manage data?
How do I manage different types of data?
How do I store and access structured data?
How do I store and access unstructured data?
Objectives
Distinguish between different types of data
Understand how to work with structured and unstructured data
Be comfortable with SQL select and filter queries
Understand what meta-data is
Managing Data
All of our research is fundamentally reliant on data so it’s important to understand the different types of data, how to store them, and how to work with them. In this lesson we’ll learn all the above!
Types of data
Fundamentally computers store data as 0s and 1s, representing floats, integers, boolean, or character data types. However this is not what we are going to talk about here. Here we are more concerned with a higher level of abstraction, which includes the following data types:
- raw data
- as it comes directly from a sensor or data source
- contains signal + noise
- format and quality is highly dependent on the data source
- clean data
- has bad/invalid records removed or flagged
- has been converted to a format which is usable by your analysis or processing tools
- transformed data
- useful features have been identified
- low level aggregation has been applied
- instrument/sensor/model dependent effects have been removed
- reduced data
- has been modeled
- has been processed into a different form such as images or a time series
- published data
- the highest level of abstraction/aggregation of your data
- often represented as tables or figures
The following describes a generic data processing workflow
Discussion
In your groups have a look at the different types of data and stages of the data processing workflow. Think about:
- Where do you get your data from?
- Are there more than one source of data?
- How do you keep track of which data is in which stage of processing?
- When you publish your work, which data products do you want to keep and which can be deleted?
Record some notes in the shared document.
We can classify data by the processing stage as was done above, but also in an orthogonal dimension of how the data is organized. The two most general considerations here are either structured or unstructured data.

The way that we can store and access data depends on how it’s structured.
Structured data
One of the easiest ways to store structured data are in one or more tables.
The tables themselves can be stored as .csv, .fits, or .vot tables which can be read by many astronomy software tools such as topcat.
Google sheets, Microsoft Excel, or LibreOffice Calc can also be beneficial for storing structured data, particularly because they allow you to do some data exploration, cleaning, filtering, and analysis within the file itself.
When your data volumes become large, or you find yourself needing to have multiple related tables, then you probably want to start exploring databases as a storage option. See the Bonus Content
Unstructured data
Unstructured data cannot be aggregated or combined into a single data format due to their lack of common structure. In such cases it is often best to make use of a regular directory/file structure to store the individual files in a way that can be navigated easily. A well planned directory/file structure it is very useful to store meta-data about each of the files that you are storing. For example you could have:
- A
README.mdfile in the root directory that describes the directory/file structure - A
metadata.csvfile that maps the location of a file to some high level information such as the file format, the date the file was collected, the processing stage of the data, or the version of the file.
Exercise
- Download example data example.zip
- Unzip the file and have a look at the content
- See if you can infer the purpose of each file
- Make a plan for how you would store and manage this data
- Note in the shared document what you have decided for at least one of the files
Data cleaning
In the previous lesson we worked with a file Australian_Meteorite_Landings.csv.
For your benefit this had already been cleaned to remove invalid entries, attributes that we didn’t need, and entries that were not relevant.
Let’s now start with the original data set and go through the cleaning process.
wget -O Meteorite_Landings.csv https://data.nasa.gov/api/views/gh4g-9sfh/rows.csv?accessType=DOWNLOAD
# or
curl -o Meteorite_Landings.csv -O https://data.nasa.gov/api/views/gh4g-9sfh/rows.csv?accessType=DOWNLOAD
Now we’ll explore the data set using pandas.
View the raw data
Using your viewer of choice, have a look at the csv file that we just downloaded. Think about and discuss:
- Are there any missing data?
- What can be done about the missing data?
- Are there any columns that are not useful?
Note in our shared document which tool you used to inspect the data, and one that you would take to clean this data.
Solution with pandas
import pandas as pd df = pd.read_csv('Meteorite_Landings.csv') print("Quick look of data") print(df) print("Summary of data") print(df.describe())Quick look of data name id nametype recclass mass (g) fall year > reclat reclong GeoLocation 0 Aachen 1 Valid L5 21.0 Fell 1880.0 50.> 77500 6.08333 (50.775, 6.08333) 1 Aarhus 2 Valid H6 720.0 Fell 1951.0 56.> 18333 10.23333 (56.18333, 10.23333) 2 Abee 6 Valid EH4 107000.0 Fell 1952.0 54.> 21667 -113.00000 (54.21667, -113.0) 3 Acapulco 10 Valid Acapulcoite 1914.0 Fell 1976.0 16.> 88333 -99.90000 (16.88333, -99.9) 4 Achiras 370 Valid L6 780.0 Fell 1902.0 -33.> 16667 -64.95000 (-33.16667, -64.95) ... ... ... ... ... ... ... ... ..> . ... ... 45711 Zillah 002 31356 Valid Eucrite 172.0 Found 1990.0 29.> 03700 17.01850 (29.037, 17.0185) 45712 Zinder 30409 Valid Pallasite, ungrouped 46.0 Found 1999.0 13.> 78333 8.96667 (13.78333, 8.96667) 45713 Zlin 30410 Valid H4 3.3 Found 1939.0 49.> 25000 17.66667 (49.25, 17.66667) 45714 Zubkovsky 31357 Valid L6 2167.0 Found 2003.0 49.> 78917 41.50460 (49.78917, 41.5046) 45715 Zulu Queen 30414 Valid L3.7 200.0 Found 1976.0 33.> 98333 -115.68333 (33.98333, -115.68333) [45716 rows x 10 columns] Summary of data id mass (g) year reclat reclong count 45716.000000 4.558500e+04 45425.000000 38401.000000 38401.000000 mean 26889.735104 1.327808e+04 1991.828817 -39.122580 61.074319 std 16860.683030 5.749889e+05 25.052766 46.378511 80.647298 min 1.000000 0.000000e+00 860.000000 -87.366670 -165.433330 25% 12688.750000 7.200000e+00 1987.000000 -76.714240 0.000000 50% 24261.500000 3.260000e+01 1998.000000 -71.500000 35.666670 75% 40656.750000 2.026000e+02 2003.000000 0.000000 157.166670 max 57458.000000 6.000000e+07 2101.000000 81.166670 354.473330
solution with bash
head Meteorite_Landings.csvname,id,nametype,recclass,mass (g),fall,year,reclat,reclong,GeoLocation Aachen,1,Valid,L5,21,Fell,1880,50.775000,6.083330,"(50.775, 6.08333)" Aarhus,2,Valid,H6,720,Fell,1951,56.183330,10.233330,"(56.18333, 10.23333)" Abee,6,Valid,EH4,107000,Fell,1952,54.216670,-113.000000,"(54.21667, -113.0)" Acapulco,10,Valid,Acapulcoite,1914,Fell,1976,16.883330,-99.900000,"(16.88333, -99.9)" Achiras,370,Valid,L6,780,Fell,1902,-33.166670,-64.950000,"(-33.16667, -64.95)" Adhi Kot,379,Valid,EH4,4239,Fell,1919,32.100000,71.800000,"(32.1, 71.8)" Adzhi-Bogdo (stone),390,Valid,LL3-6,910,Fell,1949,44.833330,95.166670,"(44.83333, 95.> 16667)" Agen,392,Valid,H5,30000,Fell,1814,44.216670,0.616670,"(44.21667, 0.61667)" Aguada,398,Valid,L6,1620,Fell,1930,-31.600000,-65.233330,"(-31.6, -65.23333)"wc Meteorite_Landings.csv45717 166894 3952161 Meteorite_Landings.csv
You may have noticed when inspecting the file above, that the header names are occasionally useful, but don’t do a good job of explaining what the data represent. For example:
- what does the “rec” in reclat/reclong represent?
- Why do some meteors have blank reclat/reclon, while others have (0,0)?
- Did someone found a meteorite at the Greenwich observatory?
- There are many meteorites with the same reclat/reclong, why is this?
- What does recclass represent?
Here is where we need to think about the meta-data attached to this table. We need context (how/why the table was created), and also a description of what the data represent. If we go to the NASA website which provides this data, they have a description of the column data. However, it’s not so helpful:
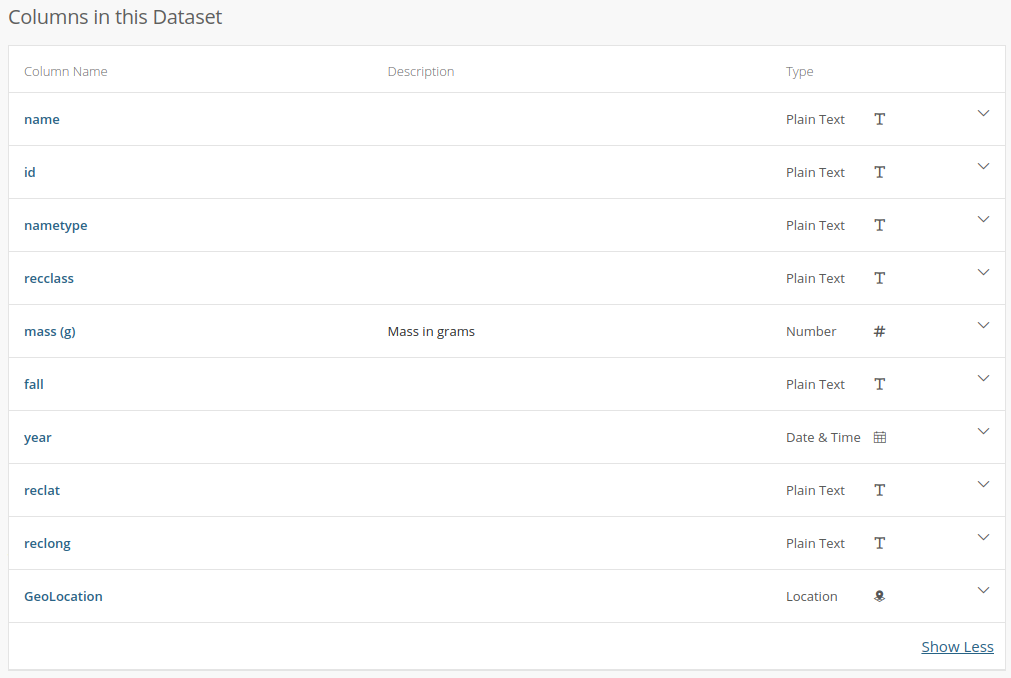
Wuh-wah! The only column with a description is the one that was most obvious. Not only that, but the data type listed for the columns is incorrect in many cases. For example, id, reclat, and reclat are listed as being “plain text” but they are clearly numeric (int, and floats), while “year” is listed as being “date and time”.
Enough complaining, lets start fixing.
Challenge
We want to plot all the Australian meteorites, with a measured mass value, and a nametype of “Valid”
- Using pandas, load the meteorite dataset
- Delete the geolocation column as it’s a duplicate of the reclat/reclong columns
- Select all rows with a non-zero and non-null mass value
- Count the rows and record this number
- Delete these rows
- Repeat the above for:
- rows with a
reclat/reclongthat are identically 0,0- rows with a blank
reclat/reclong- rows with
nametypethat is not “Valid”- Delete the
nametypecolumn as it’s now just has a single value for all rows- Choose a bounding box in lat/long for Australia (don’t forget Tassie!)
- Select, count, and delete all rows that are outside of this bounding box
- Save our new dataframe as a
.csvfile- Run our previously created script on this file and view the results
Data storage and access
The way that you store and access data depends on many things including the type of data, the structure of the data, and the ways that you will access and use the data.
We’ll focus here on the two data types introduced at the start of this lesson: structured and unstructured data.
Structured data
These are the kinds of data that we can store as rows in a .csv file or Excel spreadsheet.
The two options for storing these data would be flat files or a database.
A flat-file is, as the name suggests, a single file which contains all the information that you are storing.
You need an independent program to load/sort/update the data that is stored in a flat-file.
As the data volume becomes large, or the relation between data in different flat-files becomes more intertwined the creation, retrieval and management of these files can become time consuming and unreliable.
A database is a solution that combines the data storage, retrieval, and management in one program. A database can allow you to separate the way that the data are stored logically (tables and relations) and physically (on disks, over a network).
We’ll explore some basics of data bases using sqlite3.
This program supports the core functionality of a relational database without features such as authentication, concurrent access, or scalability.
For more advanced features you can use more full featured solutions such as PostgreSQL or MySQL.
One of the nice things about sqlite3 is that it stores all the data in a single file, which can be easily shared or backed up.
For this lesson we’ll be working with a ready made database Meteorite_Landings.db.
Using SQL to select data
There are two choices of how you can use sqlite3 for this lesson:
- run
sqlite3 Meteorite_Landings.dbfrom your terminal - Go to sqliteonline.com and upload the file
The data in a database is stored in one or more tables, each with rows and columns. To view the structure of a table we can use:
.schema
CREATE TABLE IF NOT EXISTS "landings" (
"index" INTEGER,
"name" TEXT,
"id" INTEGER,
"nametype" TEXT,
"recclass" TEXT,
"fall" TEXT,
"year" REAL,
"reclat" REAL,
"reclong" REAL,
"GeoLocation" TEXT,
"States" REAL,
"Counties" REAL,
"mass" REAL
);
What we see in the output here are the instructions on how the table was created.
The name of the table is landings, and there are a bunch of column names which we recognize from previous work.
We can see that each column has a particular data type (TEXT, INTEGER, REAL).
In a database the types of data that can be stored in a given column are fixed and strictly enforced.
If you tried to add “one” to a column with type REAL, then the database software would reject your command and not change the table.
If we want to see the content of the table, we need to SELECT some data. In SQL we can do this using the SELECT command as follows:
SELECT <columns> FROM <table>;
SELECT name, mass, year FROM landings;
SELECT * FROM landings;
The last command is the most common command for just looking at the table content as it will show all the columns without you having to list them all.
If we want to show only the first 10 lines (like head) then we can choose a LIMIT of lines using:
SELECT * FROM landings LIMIT 10;
Note that if you are using the command line version of sqlite3 the default output can be a little hard to read.
Firstly, you’ll get all the data speeding by. Secondly, the data are not well formatted for humans to read.
We can fix this with a few commands:
.mode column
.headings on
Combining all this together we have the following:
sqlite> SELECT * FROM landings LIMIT 10;
index name id nametype recclass fall year reclat reclong GeoLocation States Counties mass
---------- ---------- ---------- ---------- ---------- ---------- ---------- ---------- ---------- ----------------- ---------- ---------- ----------
0 Aachen 1 Valid L5 Fell 1880.0 50.775 6.08333 (50.775, 6.08333) 21.0
1 Aarhus 2 Valid H6 Fell 1951.0 56.18333 10.23333 (56.18333, 10.233 720.0
2 Abee 6 Valid EH4 Fell 1952.0 54.21667 -113.0 (54.21667, -113.0 107000.0
3 Acapulco 10 Valid Acapulcoit Fell 1976.0 16.88333 -99.9 (16.88333, -99.9) 1914.0
4 Achiras 370 Valid L6 Fell 1902.0 -33.16667 -64.95 (-33.16667, -64.9 780.0
5 Adhi Kot 379 Valid EH4 Fell 1919.0 32.1 71.8 (32.1, 71.8) 4239.0
6 Adzhi-Bogd 390 Valid LL3-6 Fell 1949.0 44.83333 95.16667 (44.83333, 95.166 910.0
7 Agen 392 Valid H5 Fell 1814.0 44.21667 0.61667 (44.21667, 0.6166 30000.0
8 Aguada 398 Valid L6 Fell 1930.0 -31.6 -65.23333 (-31.6, -65.23333 1620.0
9 Aguila Bla 417 Valid L Fell 1920.0 -30.86667 -64.55 (-30.86667, -64.5 1440.0
By convention we use all caps for the keywords, and lowercase for table/column names.
This is by convention only, and sqlite3 is not case-sensitive.
We can filter the data that we retrieve by using the WHERE clause.
SELECT <columns> FROM <table> WHERE <condition> LIMIT 10;
Challenge
- Find all the meteorites that fell between 1980 and 1990, and have a mass between 100-1,000g.
- List the name and reclat/reclong for 15 of these meteorites
Solution
sqlite> SELECT name, reclat, reclong FROM landings WHERE year >= 1980 AND year <=1990 LIMIT 15;name reclat reclong ---------- ---------- ---------- Akyumak 39.91667 42.81667 Aomori 40.81056 140.78556 Bawku 11.08333 -0.18333 Binningup -33.15639 115.67639 Burnwell 37.62194 -82.23722 Ceniceros 26.46667 -105.23333 Chela -3.66667 32.5 Chiang Kha 17.9 101.63333 Chisenga -10.05944 33.395 Claxton 32.1025 -81.87278 Dahmani 35.61667 8.83333 El Idrissi 34.41667 3.25 Gashua 12.85 11.03333 Glanerbrug 52.2 6.86667 Guangnan 24.1 105.0
You can interact with a database programmatically using the python module sqlite3.
With the combination of sqlite3 and pandas modules, you can save a pandas data frame directly to a database file.
This is one of the easiest ways to create a simple database file.
import sqlite3 as sql
import pandas as pd
# read the .csv file
df = pd.read_csv("Meteorite_Landings.csv")
# create a connection to our new (empty) database
con = sql.connect("Meteorite_Landings.db")
# update the name of mass column to avoid spaces and special chars
db['mass'] = db['mass (g)']
del db['mass (g)']
# save the dataframe to the database via the connection
df.to_sql(name='landings', con)
Unstructured data
Data that doesn’t conform to a standard tabular format are referred to as unstructured data. For example:
- images,
- videos,
- audio files,
- text documents
For these data we still need to be able to know where it is and where to put new data. In this case we can impose some structure using a directory/file structure. The structure that you use ultimately depends on how you’ll use the data.
For example, imagine that we have a photograph of each of the meteorites represented in our landings table from the previous exercise.
The images could be organized in one of the following ways:
name.png
<name>/<location>/id.png
<continent>/<year>/id.png
<recclass>/<year>/name.png
<state>/<county>/<year>/<recclass>/id.png
In each of the above examples you can see that there is an intentional grouping and sub-grouping of the files which make sense for a given type of analysis, but will be confusing or tedious to work with for a different type of analysis.
For example, <continent>/<year>/id.png will be convenient if you are looking at how meteorite monitoring/recovery has progressed in various countries over time, but it will be annoying to work with if you just want to select meteorites with a particular composition.
Whatever choice you make for storing your data, it is a good idea to make a note of this scheme in a file like README.md or file_structure.txt, so that you and others can easily navigate it.
You could start to encode extra information into the filenames, and then describe this naming convention in the README.md file.
A very nice hybrid solution is to use a database to store some information about your image files in a table, and have a column which is the filename/location of the image file.
In our example, this would mean appending a single new column to our existing database table or .csvfile.
In the case where the data are the images, then you would need to start extracting information about the data (meta-data) from each of the files, and then store this in a database or table.
For example if you have a set of video files you could extract the filming location (lat/long), time, duration, filename, and some keywords describing the content.
This solution allows you to query a database to find the files that match some criteria, locate them on disk, and then use some video analysis software on each.
Meta-data
- Think about some of the unstructured data that you collect or create as part of your research work
- Write a short description of what these data are
- Choose 4-5 pieces of metadata that you would store about these data
- Use our shared document to share you description and meta-data
Key Points
Invest time in cleaning and transforming your data
Meta data gives context and makes your data easier to understand
How and where you store your data is important
Afternoon Tea
Overview
Teaching: min
Exercises: minQuestions
Objectives
Time for our last break.
Key Points
Project Management
Overview
Teaching: 30 min
Exercises: 30 minQuestions
What is project management?
How can I manage my PhD?
What tools are available to help me with time and project management?
How can I use my project management skills on my supervisor?
Objectives
Understand how to use a kanban board to manage a project
Reflect on your current and future time management
Make a plan to ‘own’ your PhD project
Project management
The goal of project management is to make it easier to understand the work that needs to be done, track the progress of individual work items, to reduce the communications overhead, and to increase productivity of all involved. By applying project management strategies to your work you can expect to get more work done with your limited resources, have a clearer view of the current state of your projects, and be able to collaborate with others more effectively.
Discussion
Have you ever experienced any of the following?
- Struggling to find the email with the plot you want to share in a meeting
- Not being sure if you have the most up to date version of a document
- Struggling to keep track of your tasks
- Forgetting deadlines
- Taking on too much work so you can’t work on your project
- Struggling to keep track of who was assigned to a task
- Being unsure who is doing the task that is blocking your current work
- Having meetings that involve updates on tasks but no helpful discussions
In small groups share you experience and potential solutions to some of the above situations. Choose one issue and solution and add it to our shared document.
Project management basics
The two main project management styles that are applied in software development, business, and research are waterfall and agile. The two styles represent different ends of a flexibility spectrum.
In the waterfall mindset there is a very linear approach to the design and execution and delivery of the project with the main focus being on compliance and following a set process. This works well in situations that are very risk averse, or have a large amount of external oversight. You’ll see waterfall style project management in large projects like instrument development, construction, and commissioning.
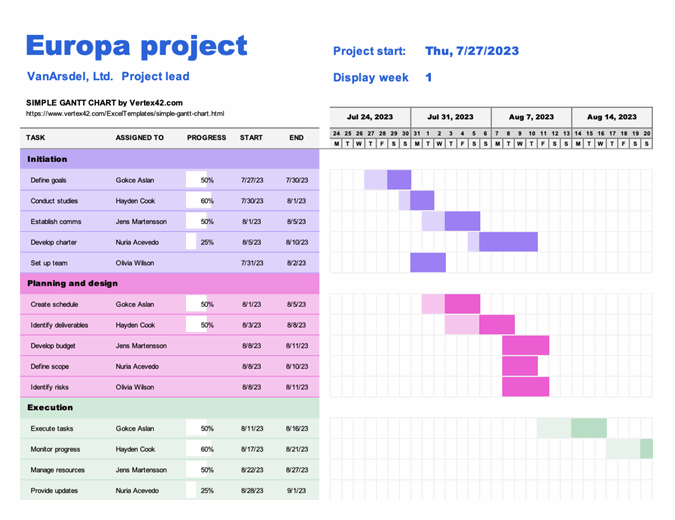
Due to the rigid design of waterfall managed projects, the work requirements are often visualised in a Gantt chart such as the one below:
 |
|---|
| The waterfall management style gets it’s name from the fact that work items are completed in a linear fashion with a cascading dependency chart such as that above. |
With a agile mindset the main focus is on outcomes and deliverables, with the design and execution and delivery occurring in cycles. An agile approach works well in situations where the path to completion is not well defined, or where resources or requirements are liable to change throughout the project. Without knowing it you will probably already be engaged in a very agile-like approach to your research projects simply because so much of your work is likely to be exploratory in nature.
 |
|---|
| In a research context, an agile approach is all about exploring and responding to change (discoveries). You don’t know the outcome of your work before you start. |
A common tool for visualizing and tracking work, particularly in an agile process, is the kanban board. The Kanban board was initially as a physical pin board on which cards or sticky notes were used to represent tasks. The tasks are organized into columns based on their status, and would typically migrate left to right across the board. Columns are customized for each project, but there are typically no more than 4-5. If a board is tracking multiple projects, or a project has multiple deliverables, then the board can also be divided horizontally into swimlanes. As the name suggests, tasks can move within a swimlanes, but are not intended to change lanes.
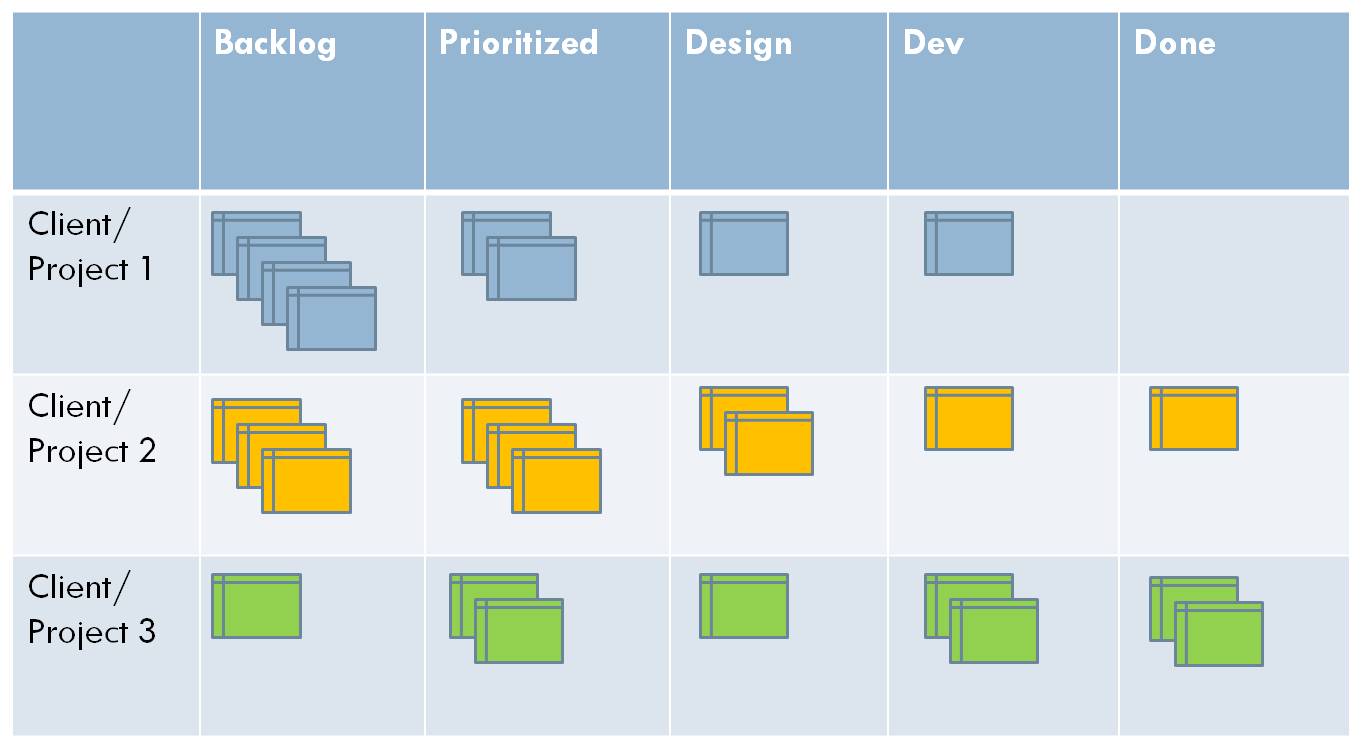 |
|---|
| An example of a Kanban board with 5 columns and 3 swimlanes |
Design a board
Think of a project that you are currently working on and design a Kanban board for it. Consider the following:
- How many task status categories are needed and what do you label them.
- Could you get away with fewer categories?
- Would it be useful to divide your tasks into different swimlanes, and if so how?
- What tasks could you put in each category right now?
Using a simple markdown format create a table in our shared document that shows the categories (and swimlanes if used) that you would use for your project. If you have time include some example tasks. (See the example table).
Online Kanban with Trello
Trello is one of the most popular project management applications thanks to it being easy to use, flexible, and free.
If you have not already, create a Trello account here.
Trello organizes your tasks by workspaces and boards. Tasks that relate to a project are grouped into the same boards, and then multiple related boards are grouped into a workspace. For example you may have a different board for each of your research projects, but then group them together based on your collaborations. If you are doing a PhD then you’ll probably just need a single board for your PhD, and a single workspace to hold that board. As an early career researcher you’ll probably have a workspace that is something like “my astro work”, which then has a different board for each of the projects that you are involved in. Many people find it useful to also have a “personal” or “home life” board that they use to track non-work related activities. These are good people.
Trello uses a Kanban board to track tasks for each of your boards, and this is the default visualization that you’ll see when you create a board. The different columns within a board are referred to as “lists” and items within the list are “cards”. You will have the freedom to create/rename/delete the lists as you see fit, and to move cards between lists as they move through the to-do/doing/done phases that you define.
Cards have a lot of features that you can play with. Some of the immediately useful ones include:
- a due date
- embedded check lists for incremental tasks
- customizable coloured labels
- attachments
- The ability to use markdown to format the text of the card
- A chat/discussion space
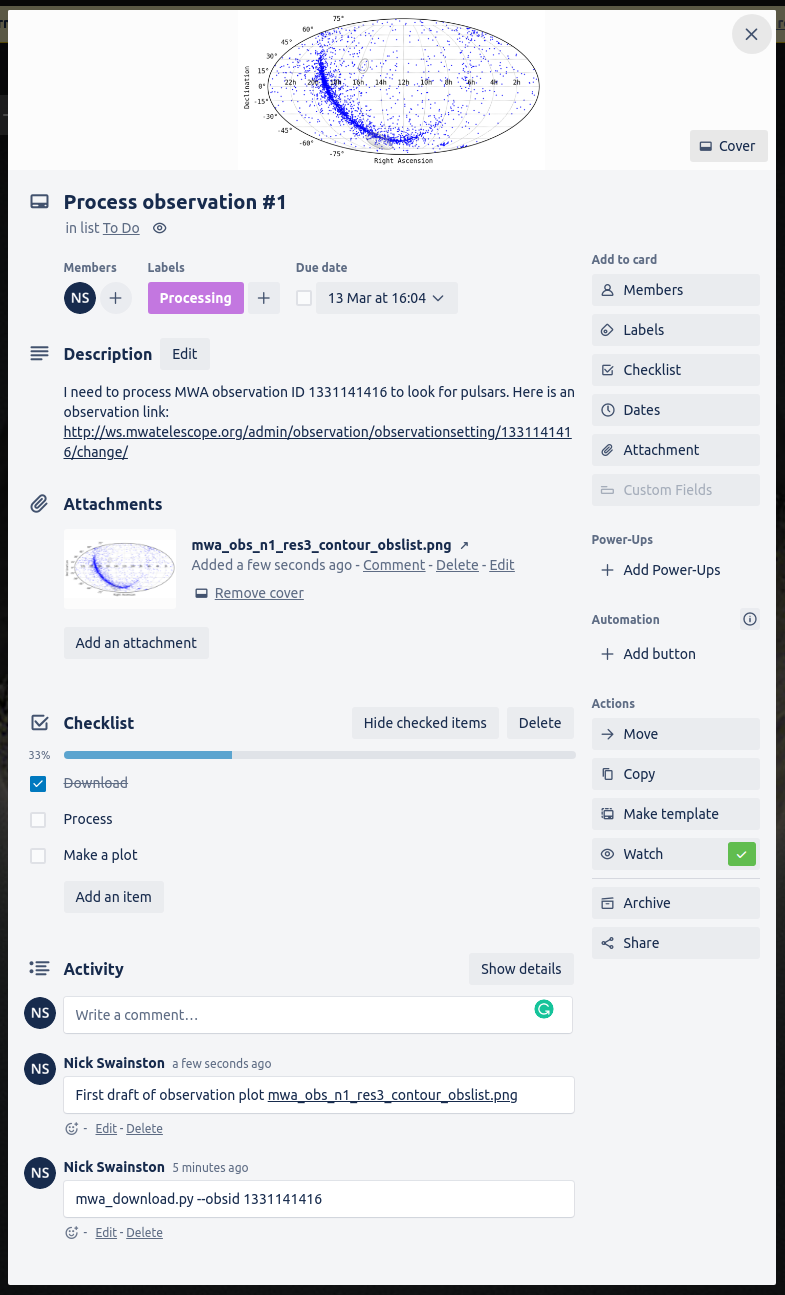 |
|---|
| An example of some of the features that you can add to your cards |
Now that we have that nomenclature out of the way, lets get started with our Trello accounts.
Create a board with some dummy cards
- Create a new workspace
- Within that workspace create a new board
- Within that board create three columns “todo”, “doing”, and “done”
- Within the “todo” column create a new card
- Set the title of the card to be something like “Create a card”
- Move the card into the “doing” column
- Add yourself as a member of the card
- Add a short description to the card
- Move the card to the “done” column
Pat yourself on the back for a job well done.
Time management
Part of managing a project is realizing that time is a very limited resource so it is often beneficial to manage it to the best of your abilities. You can split time management into two main catagories, prospective (planning) and retrospective (tracking).
A common way of estimating time projects will take is to work in hours/days/weeks/months of effort. However you are often not working on a single task at a time so it can be easier to break your total time available (your Full-Time Equivalent or FTE), into fractions, and then assign these fractions to different tasks. By estimating and tracking your FTE over the course of a project you can ensure that you are not spread too thin and you can finish tasks on time. Effective time management will prevent you from taking on more work than you can realistically complete.
For example, someone asks you to help with a project. You look at your project planning app and see that you have two projects you aim to spend 0.4 FTE on and a third you aim to spend 0.2 FTE on. Since this fills up the entire 1.0 FTE, you let them know that you can not assist on their project. You know that your third project will be complete in a month, so you offer to provide help at the level of 0.2 FTE starting in a month.
Deciding where you spend your FTE is only half the battle. You must also track your time to ensure that you are staying on budget, and adjusting if required. Tracking your time can be done through several apps (see the following section) and should give you at least a rough estimate of the time you spend on each project. This basic information will allow you to see how much time tasks and projects take and improve your future estimates. For example, you estimated that it would take you a month to write the first chapter of your thesis, but it took six weeks. You now know to budget more time for the next chapter. Another example is that you budget 0.5 FTE for teaching, but you realize you are using 0.6 FTE, so you will have to cut back on other projects.
How to track your time
There is a large variety of time tracking techniques and software available to help you track your time. Probably the most useful for academic work is an online or mobile app. Some time tracking apps are feature rich with all kinds of integrations and billing options, however they can come with a large time overhead which can be counter productive. If you are spending more than 10 minutes per day tracking your time, it is unlikely to be worth the effort, so we will focus on simple time tracking methods.
Excel method
Here is a link to a simple Google Sheets template (shown below) of an easy way to track how much time you spend per week on several projects.
At the end of each week, you update the sheet to track the fraction of your time you spent on each project.
Additional projects can be added easily as new columns.
This is effective for when you only care about your fractional FTE at a fairly low resolution.
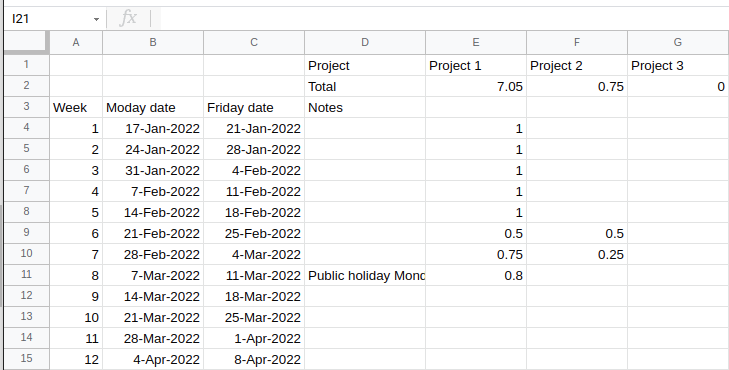
Chronos app
Chronos is a simple app that will allow you to keep track of your project time.
Simply make a new project with a descriptive name and set the billing type to non-billable.
You can start a timer for the project you are currently working on or manually add how many hours you have spent.
This method is good for when you have many projects that you are swapping between throughout the day, or when you want to be able to track your ‘effective’ work hours within a week.
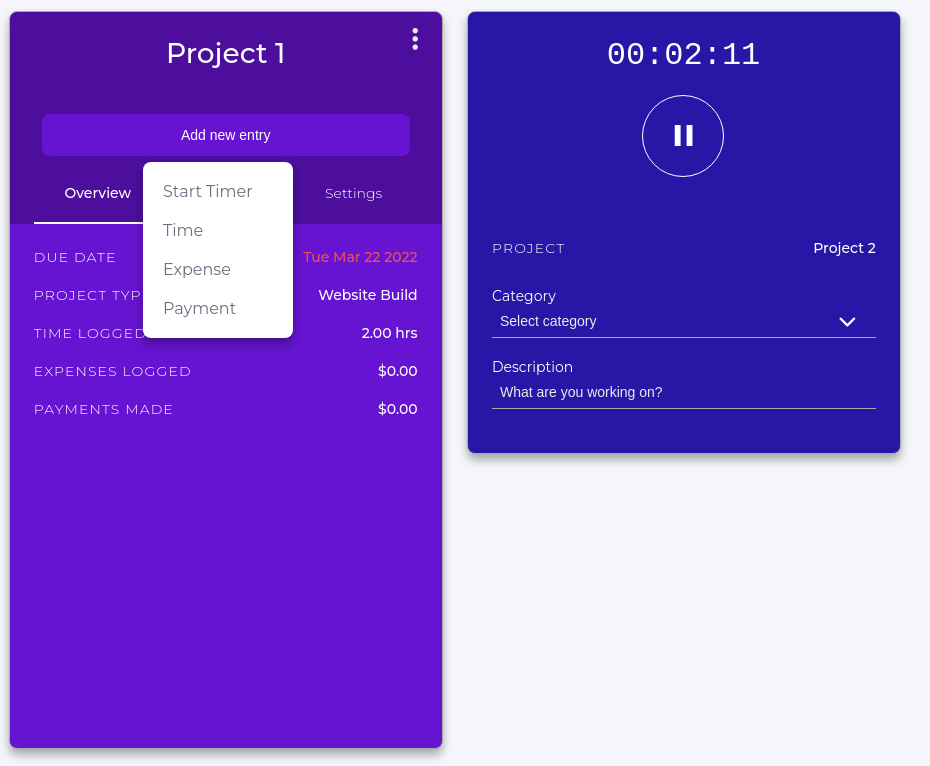
How do you spend your time?
Think about the work that you do as part of your research.
- Itemize the projects/work that you do (up to 10 max)
- For each item how much time you want to spend on it
- For each item estimate how much you think you spend on it
- Use our shared document to comment on why these last two might be different
Consider:
- Of the three methods described for time tracking which would be more useful to you
“Owning” your project
Your PhD and research career is a story in progress and you are the hero protagonist. As with any good story, the hero goes through an arc of personal development. For you, this will likely be a progression from being led by your supervisor in a project that they conceived to being the expert in an area and being the main driver of your research. You will likely start out turning up to meetings that your supervisor calls, doing what you are told, reading all the suggested readings, and relying on your supervisor for the “next steps”. Eventually you will turn up to meetings that you organize, deciding on what your project goals are and how you’ll progress toward them, and handing out todo items at the end of a meeting. This is what we call “owning” your project, and when it happens you’ll feel less like an imposter, and more in control of your work (and life!). When your supervisor or mentor sees this transition is also very rewarding for them!
Much of the above will come naturally with time, but there are things that you can start thinking about now that will help you own your project now. Some key owning behaviors include:
- Setting clear expectations for what work needs to be done, including deadlines
- Let people know how much time you have to work on something and when you expect it to be complete
- Clear communication on progress, including any risks or slippages
- Let people know when you complete tasks or if there are road blocks
- Taking the lead on project ideas
- You don’t have to have all the solutions, but making suggestions and asking the right questions is extremely helpful to everyone
- Having meeting agendas and showing up to meetings with all the information that you need
- Send plots/text ahead of time for review, have the relevant files/pages open on your laptop ready to share
Discussion - how do/will you own your project
Think about the relationship you have with your group/supervisor
- Identify an owning behavior that you are already doing
- Identify an issue that you have and would like some advice with
- Identify something that you are currently doing that has avoided some of the problems that we started the lesson with link
Share your ideas in our shared document
Key Points
Project and time management is key to success
Many tools exist, find one that works for you and stick with it
You PhD project is your’s for the taking
Bonus Content
Overview
Teaching: 0 min
Exercises: 0 minQuestions
What else is there?
Objectives
review some content that was too important to skip but didn’t fit in our lessons
Bonus Content
Bash as a scripting language
Bash is far more than a terminal, it’s a whole language in it’s own right.
What we use as our terminal is equivalent to the interpreter we interact with when we type python3 on the command line.
A few key things to know about bash to get you started are:
- variables can be set like
fname=myfile.csv(with no spaces around the=) - variables can be accessed using
${fname}or the slightly more relaxed$fname - A file that starts with
#! /usr/bin/env bashand has execute permissions (chmod +x <filename>) can be run directly - flow control uses
if ... then ... else .. fisyntax - new line continuation is accomplished by having
\as the last character on the line - converseley, multiple commands can be used on a line, separate them with
; - program output can be redirected to a file with
>(replace) or>>(append)
For more info see this wikibook for a nice intro.
Depending on the task you are trying to accomplish, a bash script can be easier to work with than a python script.
Automation with aliases
A basic form of automation is the use of aliases. The goal here is to save you a bunch of keyboard or mouse clicks. Some common examples:
# long list all files
alias ll='ls -al'
# open ds9 and press about 20 buttons to get the interface set up just the way I like
alias ds9='~/Software/ds9/ds9 -scalelims -0.2 1 -tile -cmap cubehelix0 -lock frame wcs -lock scale yes -lock colorbar yes -lock crosshair wcs'
# run stilts and topcat and not have to bother with the java invocation
alias stilts='java -jar ~/Software/topcat/topcat-full.jar -stilts'
alias topcat='java -jar ~/Software/topcat/topcat-full.jar'
# activate my favorite python3 environment
alias py3='source ~/.py3/bin/activate'
Structured data storage
CSV files are great but can be cumbersome to work with when they become large or when you have multiple files with related data. An alternative is to store your data as tables in a database, where you can make explicit links between multiple tables, and make use of the SQL language to select, filter, order, and extract the data that you need. Some commercial (but free) solutions include PostgreSQL and MySQL, but these require a fair bit of system admin to get setup and work with. A great lightweight middle-ground is SQLite, which provides the core functionality of a relational database but without all the security, authentication, and sys admin requirements of a commercial solution. See this carpentries lesson for an introduction do databases using SQLite.
Task prioritization
In most cases you will have more tasks than time available to do said tasks. You will therefore need some way of determining which tasks should be done and which can be left undone or done at a later time. A common metric to decide on the order to do tasks is the priority matrix:
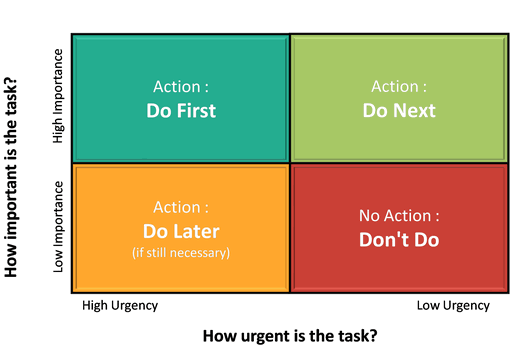
You can incorporate the above priority matrix into your work planning in a number of ways:
- Use these four categories as columns in your kanban board
- Create labels with corresponding name/colour to add to each of the tasks in your kanban board
- Create swimlanes for your kanban board that correspond to these categories (though tasks may need to change lanes!)
An alternative priority matrix which could be useful for research work is one where you consider impact (research output) and effort (time to complete). This is useful when considering which projects to take on and their goals. Successful researchers are often skilled at identifying projects that are high impact with minimal effort.
Key Points
there is always more to learn