Scientific Computing
Overview
Teaching: 30 min
Exercises: 30 minQuestions
What is scientific computing?
How can I automate common tasks?
What is reproducible research?
Objectives
Feel good about your computing goals
Appreciate various levels of automation
Understand what reproducibility means
Scientific Computing
Scientific computing is best understood by its relation to, and difference from, computer science, and software engineering. Computer science places a big focus on correctness of the code and the efficiency of the algorithm. Software engineering places a big focus on the usability of the software, and how it fits within an ecosystem of other solutions. Scientific computing cares little for any of these concerns, and is primarily concerned with producing correct scientific results. There is of course a large overlap between the three ideals as shown in the ternary diagrams below.
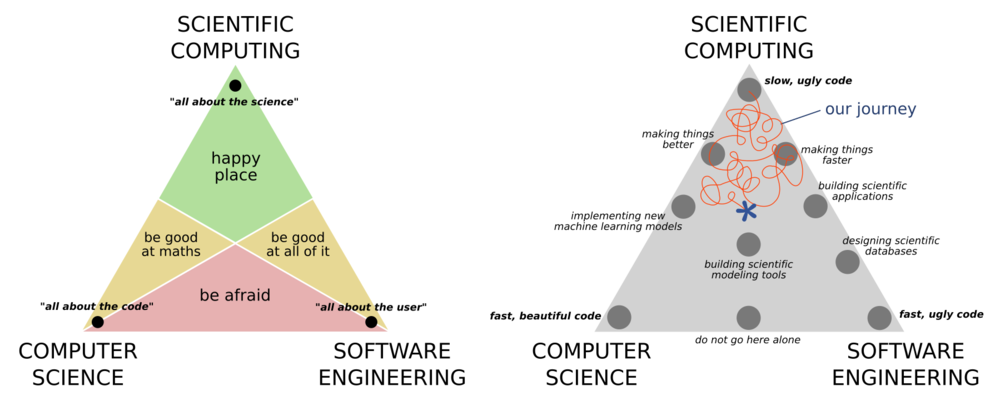
It is rare today that you can work on any project without having to some kind of computing. Many physics and astronomy students are not required to take any computing courses as part of their undergrad (though many do). Thankfully this is changing, however the courses are often focused on software engineering or computer science rather that scientific computing.
Your computing journey
In small groups, briefly share your computing journey, and where you feel that you currently sit in the above diagram.
As you discuss make notes in our shared document along the following topics:
- When writing code or developing software, what is your primary concern?
- What formal training have you had?
- What are some good resources for self/peer learning?
- What languages / tools do you commonly use or find particularly useful?
A key takeaway from the above is that you should not hold yourself to the standards of a computer scientist or a software engineer because you are neither of these things. Your goal is not to make software that is widely usable, highly performant, or scalable. First and foremost you are focusing on getting the science correct, and the rest is a bonus. Having said this, there is of course a large overlap between the three disciplines and some important lessons that we can take from software engineers and computer scientists that will make our science results more robust, more easy to replicate, easier to adapt and reuse, as well as allowing us to take on larger projects with more ambitious goals. This will be the focus of our first two lessons today.
Automate your way to success
Most of the activity in which we engage on a daily basis in research is computer based. A typical researcher will use a combination of graphical and command line user interfaces (GUIs and CLIs) to interact with this software. Each have their tradeoffs and strengths, but a major difference is that it is often easy to automate programs that have a command line interface, whilst it is difficult to do the same with a graphical interface.
Your day-to-day sciencing workflow
Think of the tasks that you do as part of your daily science-ing, and the tools that you use to make this happen. Discuss the following with your colleagues and then write in the shared document.
- Match your activity to a tool that you use, or one that you want to use.
- For each of the tools that you have identified note if it has a GUI or CLI (or both).
- What aspects of your day-to-day do you wish you could automate?
In our shared document contribute one item from each of the above.
Simple tasks are often the most easily automated. For example, maybe you want to back up the data on your (linux) computer and you have purchased an additional hard drive for this very purpose. Due to previous bad experience with RAIDed hard drives, an aversion to over complicated software solutions, and no interest in offsite or incremental backups, you have decided that your backup and recovery will consist of copying files from one hard drive to another on a regular basis. For this task you put a reminder in your calendar every monday to execute the following code:
rsync -av --delete /data/working/${USER} /data/backup/${USER}
This will use the rsync program to copy data from the hard drive mounted at /data/working/${USER} to the one mounted at /data/backup/${USER}.
Here ${USER} is the bash variable which refers to your username.
The options -av will preserve the file ownership, permissions, and access date/time, and --delete will make sure that files that are in the working directory but not in the backup directory will be deleted.
Essentially this will create an exact mirror of one drive onto another.
The program rsync is smart enough to do this mirroring by only copying the changes from one drive to another so that you don’t have to delete and re-copy the data every week.
Challenge
- What are some of the draw backs in using this method of backup?
- How could you ensure that you use the same command each time you do your backup?
- How could you automate this task so that you no longer have to think about it or spend time on it?
Solution notes
- If you don’t have your calendar open you might forget to run the program.
- You might not run the program on every Monday because you don’t work every Monday.
- Keep the above text in a file called
backup_instructions.txtso that you remember how to do it correctly.- Convert
backup_instructions.txtintodo_backup.sh, add a#!(shebang) line, and make the file executable, so you can type./do_backup.sh- You could automate by adding the above line to your crontab so that it will always run on a Monday morning without your input
Any work that you can do from the command line can be automated in a similar way. A good approach to take when considering automating a task is:
- Complete the task manually, using the command line
- Copy all the commands that you used into a file so that you have an exact record
- Save the file as a
.sh(OSX/Linux) or.batfile (Windows) - You now have an automation script!
- Next time you need to do said task, just run the script
Optional extras to make your automation even better include:
- Adding some command line arguments to your script so that you can do slightly different things each time
- Generalize the script so that information that may need to change between runs can be automatically generated or calculated
The Linux ecosystem is built on the idea of having a large number of tools each with a very specific purpose. Each tool should do one thing and do it well. There are many seemingly simple tools that already exist on your Linux (and OSX, or WSL) computer that you can chain together to great effect.
Favorite tools
- Discuss with your group some of the command line tools that you use most often.
- Let people know what tool you wish existed (because it might already!).
- Share your thoughts in our shared document.
- Have a look at the with list of others and see if you can propose a solution or partial solution.
Let’s explore some of these useful command line tools that you might use to provide some low-level automation to your life.
grep
wget
curl
sed
cut
wc
bc
We’ll explore these together, but if at a later point you want more help on these tools you can:
- use program help via the
--helpflag - use manual pages like this
man grep,man cut, or evenman man - ask dr google
Reproducible research
Reproducibility is the idea of being able to re-run your analysis with the same data and get the same results. Related concepts are replicability, robustness, and generalisabiltiy, as demonstrated in the following diagram.
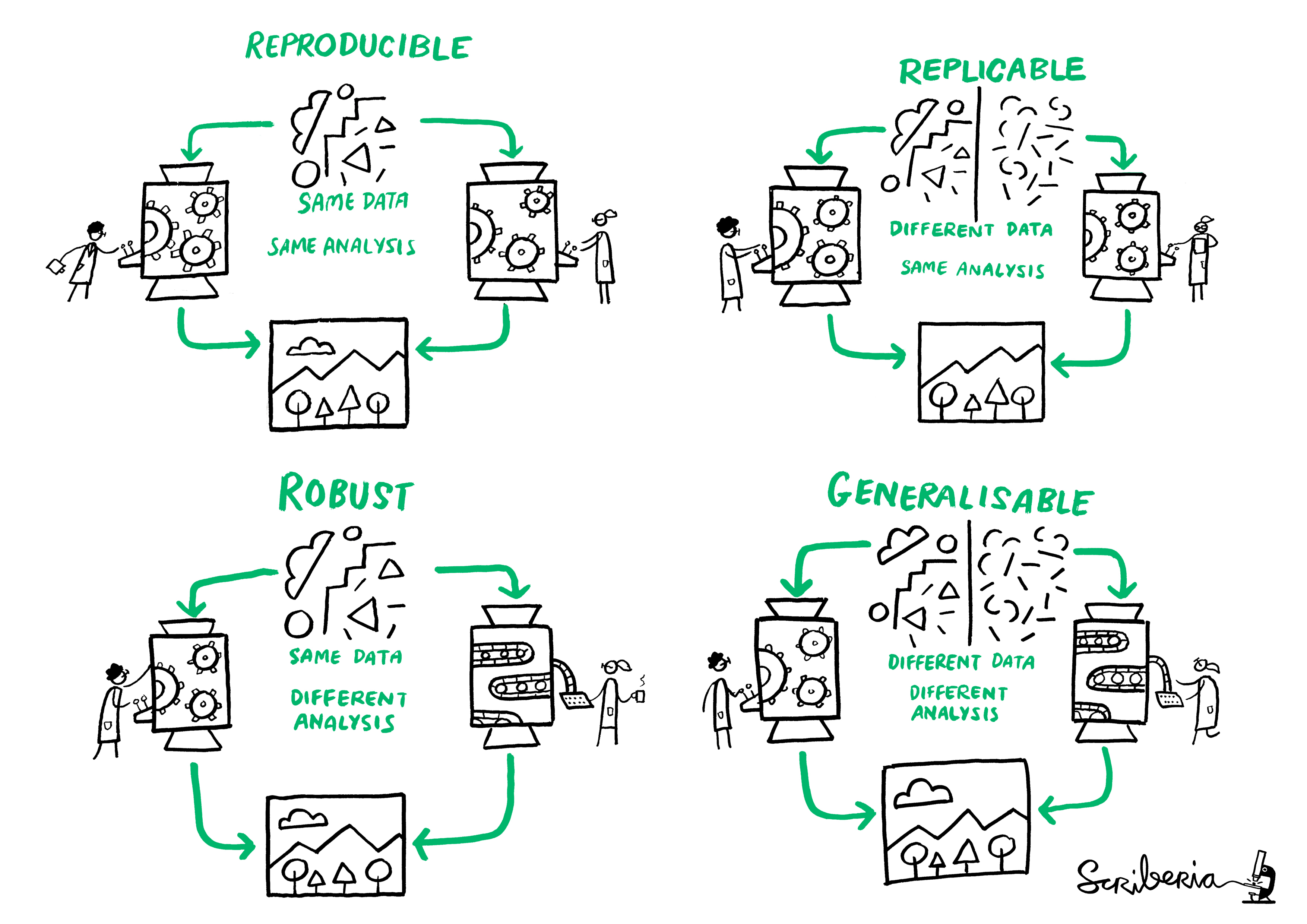
There are many reasons why you would re-run your analysis or data processing:
- As you develop your data collection, processing, and analysis techniques you will make changes to the process. Each change means re-doing all the work again.
- Your work may be extended or expanded as new data become available, requiring you to apply the same processing to different data,
- During the publication review process you need to make adjustments to the data selection or processing based on collaborator or referee’s comments,
- Other people may want to apply your techniques to their data and compare results.
A science workflow that can be consistently re-run and easily adapted is of huge benefit to you, the researcher. Sharing your data, and your methodology (via code and documentation) with others can also benefit the community because it will allow your work to be rerun and adapted, saving others time whilst generating citations and accolades for you.
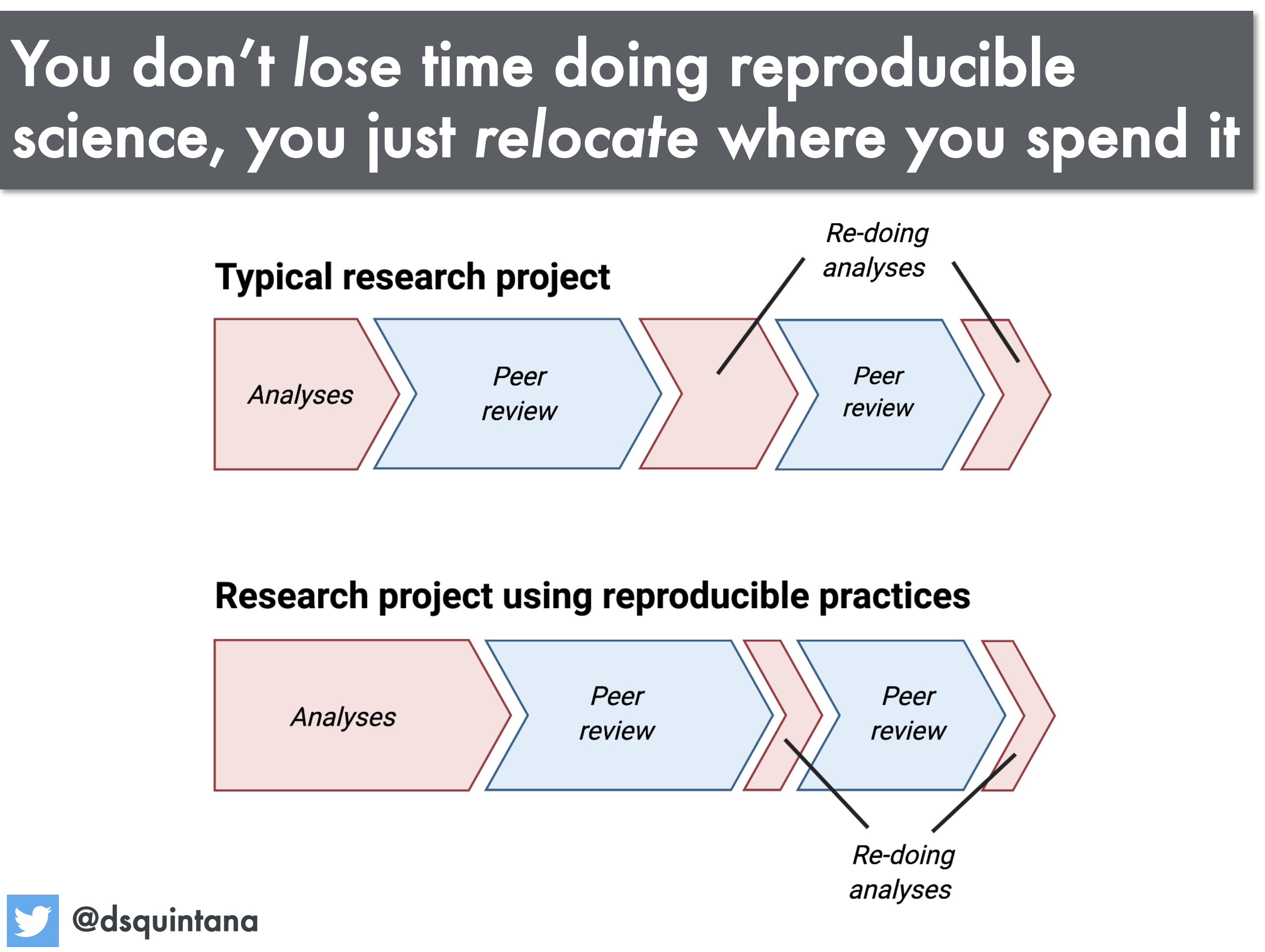
Discussion
- If you were to redo all your data processing from scratch, how confident are you that you would get the same result?
- What aspects of your workflow have you automated?
- What parts of your workflow cannot be automated, and what can you do about that?
- Is automation essential for reproducible research?
As always, share your thoughts with your group and then put some notes into the shared document.
Key Points
Scientific computing has different goals from computer science and software engineering
You are a scientist, your focus is on the science
Automation can occur at many levels
Reproducibility is good for everyone (especially you)