The Pandas module
Overview
Teaching: 45 min
Exercises: 45 minQuestions
What is Pandas?
How can I use Pandas for data exploration and visualisation?
How can I work with multiple tables at once?
Objectives
Learn about the Pandas module and some alternatives
Understand how to group and summarize data
Be able to plot with Pandas
Combine data frames sequentially or with joins
For a longer and more complete version of what will be covered in this session, please refer to lessons 3-6 of this software carpentry course.
What is Pandas?
The Python data analysis library (Pandas), is designed to be a fast, powerful, flexible, and easy to use data analysis tool built for Python. Pandas is NumFOCUS sponsored project (as are Numpy, Scipy, Matplotlib, and Astropy), and will therefore tend to “play nice” with these other packages. The Pandas library provides data structures, produces high quality plots with Matplotlib and integrates nicely with other libraries that use NumPy arrays. There are additionally many packages that are built on top of Pandas to address specialized needs (see ecosystem).
Pandas is a great solution for many of your data analysis and visualization needs however it is designed for tabular data. Alternatives to Pandas include:
| module | notes |
|---|---|
| Numpy | ndarrays can have up to 32 dimensions, but need to be of a single data type |
| Vaex | DataFrames like Pandas, with lazy evaluation of billions of rows per second. Desiged for big data. Uses memory mapping. |
| Dask | Focus on scalability and portability across architectures (laptop/desktop/cloud/HPC). Lazy evaluation, and workflow management. |
| Xarray | For working with labelled multi-dimensional arrays. Like FITS but actually good! |
Pandas provides a few different data structures that are useful for this lesson with the main one being the DataFrame. A DataFrame is a 2-dimensional data structure that can store data in columns. Each column can have a different data type, but, just like tables in a database, the data within a single column must have a consistent type. A DataFrame always has an index (0-based) for rows.
Our data
We will be using files from the Portal Project Teaching Database.
Normally this lesson would have you download the surveys.csv file from here, however we just learned about databses, so we’ll instead download the file portal_mammals.sqlite from this link.
Exploring our data
Explore our database
Open our
portal_mammals.sqlitefiles with sqlite3 or upload it to sqliteonline.View the schema of the data base by looking at the columns and data types for each of the three tables.
There are no explicit foreign key constraints on this database, however, should be able to identify which columns in the
surveystable link to each of theplotsandspeciestables.
We now connect to the data base and read it into a Pandas DataFrame:
# import our modules
import sqlite3
import pandas as pd
# connect to the database
con = sqlite3.connect('portal_mammals.sqlite')
# execute a query and save the results to a dataframe
surveys_df = pd.read_sql("SELECT * FROM surveys", con)
Pandas is aware of the environment it’s being run in, so will give slightly different out put when you are running via python3, ipython3, or a jupyter notebook.
Jupyter will give the nicest output and it will be more interactive than the iPython note book, while the output you get from a python script will be just text.
Use whatever you prefer.
If you execute the above code you should see the following output:
record_id month day year plot_id species_id sex hindfoot_length weight
0 1 7 16 1977 2 NL M 32.0 NaN
1 2 7 16 1977 3 NL M 33.0 NaN
2 3 7 16 1977 2 DM F 37.0 NaN
3 4 7 16 1977 7 DM M 36.0 NaN
4 5 7 16 1977 3 DM M 35.0 NaN
... ... ... ... ... ... ... ... ... ...
35544 35545 12 31 2002 15 AH NaN NaN NaN
35545 35546 12 31 2002 15 AH NaN NaN NaN
35546 35547 12 31 2002 10 RM F 15.0 14.0
35547 35548 12 31 2002 7 DO M 36.0 51.0
35548 35549 12 31 2002 5 NaN NaN NaN NaN
[35549 rows x 9 columns]
We can see that there were 35,549 rows parsed. Each row has 9 columns. The first column is the index of the DataFrame. The index is used to identify the position of the data, but it is not an actual column of the DataFrame (it was not the database which we read). The ellipsis (…) tell you that Pandas is intentionally not showing all the output, but just the head/tail of the table. If we have many columns (or a narrower screen) then Pandas will skip columns as well.
You can also use surveys_df.head() to view only the first few rows of the dataset in an output that is easier to fit in one window.
After doing this, you can see that pandas has neatly formatted the data to fit our screen:
surveys_df.head() # The head() method displays the first several lines of a file. It
# is discussed below.
record_id month day year plot_id species_id sex hindfoot_length \
5 6 7 16 1977 1 PF M 14.0
6 7 7 16 1977 2 PE F NaN
7 8 7 16 1977 1 DM M 37.0
8 9 7 16 1977 1 DM F 34.0
9 10 7 16 1977 6 PF F 20.0
weight
5 NaN
6 NaN
7 NaN
8 NaN
9 NaN
Exploring Our Species Survey Data
What kind of things does surveys_df contain?
DataFrames have an attribute called dtypes that answers this:
surveys_df.dtypes
record_id int64
month int64
day int64
year int64
plot_id int64
species_id object
sex object
hindfoot_length float64
weight float64
dtype: object
All the values in a single column have the same type.
For example, values in the month column have type int64.
Cells in the month column cannot have fractional values, but values in weight and hindfoot_length columns can, because they have type float64.
The object type doesn’t have a very helpful name, but in this case it represents strings (such as ‘M’ and ‘F’ in the case of sex).
Useful Ways to View DataFrame Objects in Python
There are many ways to summarize and access the data stored in DataFrames, using attributes and methods provided by the DataFrame object.
Attributes are features of an object. For example, the shape attribute will output
the size (the number of rows and columns) of an object. To access an attribute,
use the DataFrame object name followed by the attribute name df_object.attribute.
For example, using the DataFrame surveys_df and attribute columns, an index
of all the column names in the DataFrame can be accessed with surveys_df.columns.
Methods are like functions, but they only work on particular kinds of objects. As
an example, the head() method works on DataFrames. Methods are called in a
similar fashion to attributes, using the syntax df_object.method(). Using
surveys_df.head() gets the first few rows in the DataFrame surveys_df
using the head() method. With a method, we can supply extra information
in the parentheses to control behaviour.
Let’s look at the data using these.
Challenge - DataFrames
Using our DataFrame
surveys_df, try out the attributes & methods below to see what they return.
surveys_df.columns
surveys_df.shapeTake note of the output ofshape- what format does it return the shape of the DataFrame in?
surveys_df.head()Also, what doessurveys_df.head(15)do?
surveys_df.tail()Solution
surveys_df.columnsprovides the names of the columns in the DataFrame.surveys_df.shapeprovides the dimensions of the DataFrame as a tuple in(r,c)format, whereris the number of rows andcthe number of columns.surveys_df.head()returns the first 5 lines of the DataFrame, annotated with column and row labels. Adding an integer as an argument to the function specifies the number of lines to display from the top of the DataFrame, e.g.surveys_df.head(15)will return the first 15 lines.surveys_df.tail()will display the last 5 lines, and behaves similarly to thehead()method.
Calculating Statistics From Data In A Pandas DataFrame
We’ve read our data into Python. Next, let’s perform some quick summary statistics to learn more about the data that we’re working with. We might want to know how many animals were collected in each site, or how many of each species were caught. We can perform summary stats quickly using groups. But first we need to figure out what we want to group by.
Let’s begin by exploring our data:
# Look at the column names
surveys_df.columns
which returns:
Index(['record_id', 'month', 'day', 'year', 'plot_id', 'species_id', 'sex',
'hindfoot_length', 'weight'],
dtype='object')
Let’s get a list of all the species.
The pd.unique function tells us all of the unique values in the species_id column.
pd.unique(surveys_df['species_id'])
# OR
surveys_df['species_id'].unique()
which returns:
array(['NL', 'DM', 'PF', 'PE', 'DS', 'PP', 'SH', 'OT', 'DO', 'OX', 'SS',
'OL', 'RM', None, 'SA', 'PM', 'AH', 'DX', 'AB', 'CB', 'CM', 'CQ',
'RF', 'PC', 'PG', 'PH', 'PU', 'CV', 'UR', 'UP', 'ZL', 'UL', 'CS',
'SC', 'BA', 'SF', 'RO', 'AS', 'SO', 'PI', 'ST', 'CU', 'SU', 'RX',
'PB', 'PL', 'PX', 'CT', 'US'], dtype=object)
Challenge - Statistics
Create a list of unique site IDs (“plot_id”) found in the surveys data. Call it
site_names. How many unique sites are there in the data? How many unique species are in the data?What is the difference between
len(site_names)andsurveys_df['plot_id'].nunique()?Solution
site_names = pd.unique(surveys_df["plot_id"])
- How many unique sites are in the data?
site_names.sizeorlen(site_names)provide the answer: 24- How many unique species are in the data?
len(pd.unique(surveys_df["species_id"]))tells us there are 49 specieslen(site_names)andsurveys_df['plot_id'].nunique()both provide the same output: they are alternative ways of getting the unique values. Thenuniquemethod combines the count and unique value extraction, and can help avoid the creation of intermediate variables likesite_names.Note: the
.unique()function will return all the unique values includingNoneorNaNif they exist, however the.nunique()function will only count the not blank entries, so you may get a difference in counts of 1.
Groups in Pandas
We often want to calculate summary statistics grouped by subsets or attributes within fields of our data. For example, we might want to calculate the average weight of all individuals per site.
We can calculate basic statistics for all records in a single column using the syntax below:
surveys_df['weight'].describe()
gives output
count 32283.000000
mean 42.672428
std 36.631259
min 4.000000
25% 20.000000
50% 37.000000
75% 48.000000
max 280.000000
Name: weight, dtype: float64
We can also extract one specific metric if we wish:
surveys_df['weight'].min()
surveys_df['weight'].max()
surveys_df['weight'].mean()
surveys_df['weight'].std()
surveys_df['weight'].count()
But if we want to summarize by one or more variables, for example sex, we can use Pandas’ .groupby method.
Once we’ve created a groupby DataFrame, we can quickly calculate summary statistics by a group of our choice.
# Group data by sex
grouped_data = surveys_df.groupby('sex')
The pandas function describe will return descriptive stats including: mean, median, max, min, std and count for a particular column in the data.
Pandas’ describe function will only return summary values for columns containing numeric data.
# Summary statistics for all numeric columns by sex
grouped_data.describe()
# Provide the mean for each numeric column by sex
grouped_data.mean(numeric_only=True)
grouped_data.mean(numeric_only=True) OUTPUT:
record_id month day year plot_id \
sex
F 18036.412046 6.583047 16.007138 1990.644997 11.440854
M 17754.835601 6.392668 16.184286 1990.480401 11.098282
hindfoot_length weight
sex
F 28.836780 42.170555
M 29.709578 42.995379
The groupby command is powerful in that it allows us to quickly generate
summary stats.
Challenge - Summary Data
- How many recorded individuals are female
Fand how many maleM?- What happens when you group by two columns using the following syntax and then calculate mean values?
grouped_data2 = surveys_df.groupby(['plot_id', 'sex'])grouped_data2.mean(numeric_only=True)
- Summarize weight values for each site in your data. HINT: you can use the following syntax to only create summary statistics for one column in your data.
by_site['weight'].describe()Solution
- The first column of output from
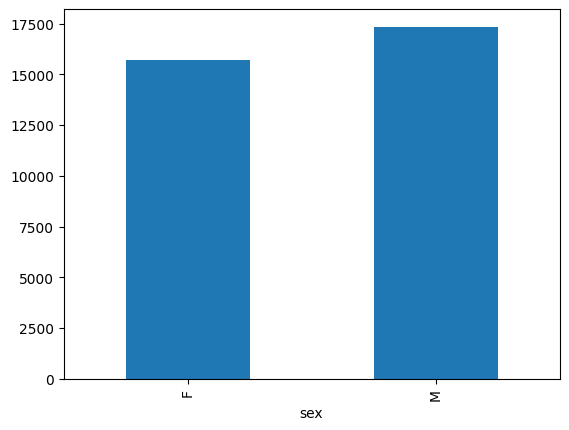
grouped_data.describe()(count) tells us that the data contains 15690 records for female individuals and 17348 records for male individuals.
- Note that these two numbers do not sum to 35549, the total number of rows we know to be in the
surveys_dfDataFrame. Why do you think some records were excluded from the grouping?- Calling the
mean()method on data grouped by these two columns calculates and returns the mean value for each combination of plot and sex.
- Note that the mean is not meaningful for some variables, e.g. day, month, and year. You can specify particular columns and particular summary statistics using the
agg()method (short for aggregate), e.g. to obtain the last survey year, median foot-length and mean weight for each plot/sex combination:surveys_df.groupby(['plot_id', 'sex']).agg({"year": 'max', "hindfoot_length": 'median', "weight": 'mean'})
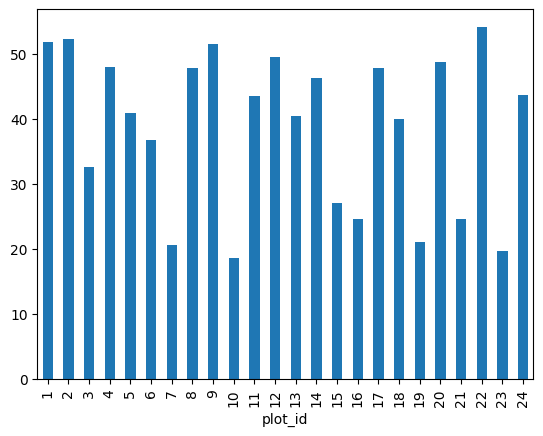
surveys_df.groupby(['plot_id'])['weight'].describe()count mean std min 25% 50% 75% max plot_id 1 1903.0 51.822911 38.176670 4.0 30.0 44.0 53.0 231.0 2 2074.0 52.251688 46.503602 5.0 24.0 41.0 50.0 278.0 3 1710.0 32.654386 35.641630 4.0 14.0 23.0 36.0 250.0 4 1866.0 47.928189 32.886598 4.0 30.0 43.0 50.0 200.0 5 1092.0 40.947802 34.086616 5.0 21.0 37.0 48.0 248.0 6 1463.0 36.738893 30.648310 5.0 18.0 30.0 45.0 243.0 7 638.0 20.663009 21.315325 4.0 11.0 17.0 23.0 235.0 8 1781.0 47.758001 33.192194 5.0 26.0 44.0 51.0 178.0 9 1811.0 51.432358 33.724726 6.0 36.0 45.0 50.0 275.0 10 279.0 18.541219 20.290806 4.0 10.0 12.0 21.0 237.0 11 1793.0 43.451757 28.975514 5.0 26.0 42.0 48.0 212.0 12 2219.0 49.496169 41.630035 6.0 26.0 42.0 50.0 280.0 13 1371.0 40.445660 34.042767 5.0 20.5 33.0 45.0 241.0 14 1728.0 46.277199 27.570389 5.0 36.0 44.0 49.0 222.0 15 869.0 27.042578 35.178142 4.0 11.0 18.0 26.0 259.0 16 480.0 24.585417 17.682334 4.0 12.0 20.0 34.0 158.0 17 1893.0 47.889593 35.802399 4.0 27.0 42.0 50.0 216.0 18 1351.0 40.005922 38.480856 5.0 17.5 30.0 44.0 256.0 19 1084.0 21.105166 13.269840 4.0 11.0 19.0 27.0 139.0 20 1222.0 48.665303 50.111539 5.0 17.0 31.0 47.0 223.0 21 1029.0 24.627794 21.199819 4.0 10.0 22.0 31.0 190.0 22 1298.0 54.146379 38.743967 5.0 29.0 42.0 54.0 212.0 23 369.0 19.634146 18.382678 4.0 10.0 14.0 23.0 199.0 24 960.0 43.679167 45.936588 4.0 19.0 27.5 45.0 251.0
Quickly Creating Summary Counts in Pandas
Let’s next count the number of samples for each species.
We can do this in a few ways, but we’ll use groupby combined with a count() method.
# Count the number of samples by species
species_counts = surveys_df.groupby('species_id')['record_id'].count()
print(species_counts)
Or, we can also count just the rows that have the species “DO”:
surveys_df.groupby('species_id')['record_id'].count()['DO']
Challenge - Make a list
What’s another way to create a list of species and associated
countof the records in the data? Hint: you can performcount,min, etc. functions on groupby DataFrames in the same way you can perform them on regular DataFrames.Solution
As well as calling
count()on therecord_idcolumn of the grouped DataFrame as above, an equivalent result can be obtained by extractingrecord_idfrom the result ofcount()called directly on the grouped DataFrame:surveys_df.groupby('species_id').count()['record_id']species_id AB 303 AH 437 AS 2 BA 46 CB 50 CM 13 CQ 16 CS 1 CT 1 CU 1 CV 1 DM 10596 DO 3027 DS 2504 DX 40 NL 1252 OL 1006 OT 2249 OX 12 PB 2891 PC 39 PE 1299 PF 1597 PG 8 PH 32 PI 9 PL 36 PM 899 PP 3123 PU 5 PX 6 RF 75 RM 2609 RO 8 RX 2 SA 75 SC 1 SF 43 SH 147 SO 43 SS 248 ST 1 SU 5 UL 4 UP 8 UR 10 US 4 ZL 2
Quick & Easy Plotting Data Using Pandas
We can plot our summary stats using Pandas, too.
# Make sure figures appear inline in Jupyter Notebook
%matplotlib inline
# Create a quick bar chart
species_counts.plot(kind='bar');
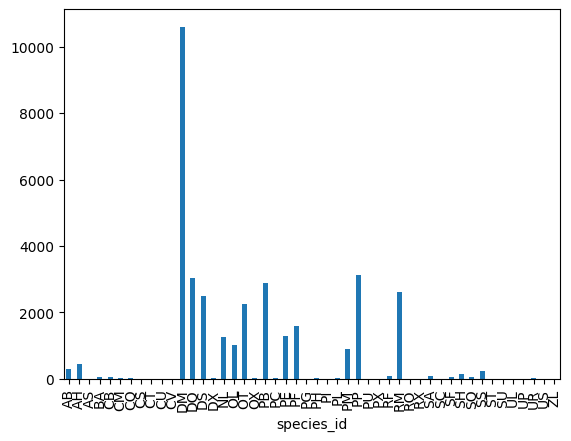 Count per species site
Count per species site
We can also look at how many animals were captured in each site:
total_count = surveys_df.groupby('plot_id')['record_id'].nunique()
# Let's plot that too
total_count.plot(kind='bar');
Challenge - Plots
- Create a plot of average weight across all species per site.
- Create a plot of total males versus total females for the entire dataset.
Solution
surveys_df.groupby('plot_id')["weight"].mean().plot(kind='bar')
surveys_df.groupby('sex').count()["record_id"].plot(kind='bar')
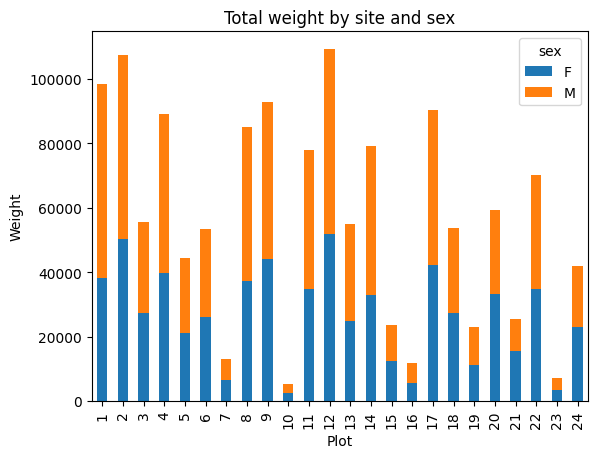
Multiple grouping
Pandas doesn’t limit us to one set of groups. For example we could group our data by plot_id and by sex:
by_site_sex = surveys_df.groupby(['plot_id', 'sex'])
site_sex_count = by_site_sex['weight'].sum()
This calculates the sums of weights for each sex within each site as a table:
site sex
plot_id sex
1 F 38253
M 59979
2 F 50144
M 57250
3 F 27251
M 28253
4 F 39796
M 49377
<other sites removed for brevity>
This isn’t the easiest to visualize, since what we really want is table with plot_id as rows, and sex as columns.
We can achieve this pivot operation by using the .unstack() function:
by_site_sex = surveys_df.groupby(['plot_id', 'sex'])
site_sex_count = by_site_sex['weight'].sum()
site_sex_count.unstack()
The unstack method above will display the following output:
sex F M
plot_id
1 38253 59979
2 50144 57250
3 27251 28253
4 39796 49377
<other sites removed for brevity>
Rather than display it as a table, we can plot the above data by stacking the values of each sex as follows:
by_site_sex = surveys_df.groupby(['plot_id', 'sex'])
site_sex_count = by_site_sex['weight'].sum()
spc = site_sex_count.unstack()
s_plot = spc.plot(kind='bar', stacked=True, title="Total weight by site and sex")
s_plot.set_ylabel("Weight")
s_plot.set_xlabel("Plot")
Combining DataFrames
There are two main ways to combine DatFrames: concatenation or by joining. To demonstrate the combining DataFrames we need an additional DataFrame to work with so we’ll load that now:
# our original surveys table
surveys_df = pd.read_sql("SELECT * FROM surveys", con)
# an additional table of species (but only some of them)
species_df = pd.read_sql("SELECT * FROM species LIMIT 20", con)
# look at our new DataFrame
species_df
species_id genus species taxa
0 AB Amphispiza bilineata Bird
1 AH Ammospermophilus harrisi Rodent
2 AS Ammodramus savannarum Bird
3 BA Baiomys taylori Rodent
4 CB Campylorhynchus brunneicapillus Bird
.. ... ... ... ...
16 GS Gambelia silus Reptile
17 NL Neotoma albigula Rodent
18 NX Neotoma sp. Rodent
19 OL Onychomys leucogaster Rodent
[54 rows x 4 columns]
Concatenation
Concatenation works by copying one DataFrame either below (vertical stack) or beside (horizontal stack) another.
We can use the concat function in pandas to append either columns or rows from one DataFrame to another.
Let’s grab two subsets of our data to see how this works.
# Read in first 10 lines of surveys table
survey_sub = surveys_df.head(10)
# Grab the last 10 rows
survey_sub_last10 = surveys_df.tail(10)
# Reset the index values to the second DataFrame appends properly
survey_sub_last10 = survey_sub_last10.reset_index(drop=True)
# drop=True option avoids adding new index column with old index values
When we concatenate DataFrames, we need to specify the axis.
axis=0 tells pandas to stack the second DataFrame UNDER the first one.
Pandas will automatically detect whether the column names are the same and will stack accordingly.
axis=1 will stack the columns in the second DataFrame to the RIGHT of the first DataFrame.
To stack the data vertically, we need to make sure we have the same columns and associated column format in both datasets.
When we stack horizontally, we want to make sure what we are doing makes sense (i.e. the data are related in some way).
# Stack the DataFrames on top of each other
vertical_stack = pd.concat([survey_sub, survey_sub_last10], axis=0)
# Place the DataFrames side by side
horizontal_stack = pd.concat([survey_sub, survey_sub_last10], axis=1)
Row Index Values and Concat
Have a look at the vertical_stack DataFrame. Notice anything unusual?
The row indexes for the two DataFrames survey_sub and survey_sub_last10 have been repeated.
We can reindex the new DataFrame using the reset_index() method.
Joining DataFrames
Pandas lets us join DataFrames as if they were tables in a database much like we did with SQL.
For example, the species table that we’ve been working with is a lookup table.
This table contains the genus, species and taxa code for 55 species.
The species code is unique for each line.
These species are identified in our survey data as well using the unique species code.
Rather than adding three more columns for the genus, species and taxa to each of the 35,549 line survey DataFrame, we can maintain the shorter table with the species information.
When we want to access that information, we can create a query that joins the additional columns of information to the survey DataFrame.
Identifying join keys
When we read our database tables into Pandas any foreign key links are lost. Instead we end up with matching values in the respective tables. This means that we can’t do the NATURAL joins that we did with sqlite, and have to be specific about how the rows should be matched up.
There are three main types of joins we can do:
- Inner join: Returns only rows which match in both tables
- Left (outer) join: Returns all rows from the first table, plus their matches in the second table.
- Will result in many columns of NULL where there is no match in the second table.
- Right (outer) join: Returns all rows from the second table, plus their matches in the first table.
- Will result in many columns of NULL where there is no match in the first table.
Inner joins
The most common type of join is called an inner join. An inner join combines two DataFrames based on a join key and returns a new DataFrame that contains only those rows that have matching values in both of the original DataFrames.
The pandas function for performing joins is called merge and an Inner join is the default option:
merged_inner = pd.merge(left=survey_sub, right=species_df, left_on='species_id', right_on='species_id')
merged_inner
In this case, species_id is the only column name in both DataFrames, so if we skipped the left_on and right_on arguments, pandas would guess that we wanted to use that column to join.
However, it is usually better to be explicit.
record_id month day year plot_id species_id sex hindfoot_length \
0 1 7 16 1977 2 NL M 32
1 2 7 16 1977 3 NL M 33
2 3 7 16 1977 2 DM F 37
3 4 7 16 1977 7 DM M 36
4 5 7 16 1977 3 DM M 35
5 8 7 16 1977 1 DM M 37
6 9 7 16 1977 1 DM F 34
weight genus species taxa
0 NaN Neotoma albigula Rodent
1 NaN Neotoma albigula Rodent
2 NaN Dipodomys merriami Rodent
3 NaN Dipodomys merriami Rodent
4 NaN Dipodomys merriami Rodent
5 NaN Dipodomys merriami Rodent
6 NaN Dipodomys merriami Rodent
The result of an inner join of survey_sub and species_df is a new DataFrame that contains the combined set of columns from survey_sub and species_df.
It only contains rows that have two-letter species codes that are the same in both the survey_sub and species_df DataFrames.
In other words, if a row in survey_sub has a value of species_id that does not appear in the species_id column of species, it will not be included in the DataFrame returned by an inner join.
Similarly, if a row in species_df has a value of species_id that does not appear in the species_id column of survey_sub, that row will not be included in the DataFrame returned by an inner join.
The two DataFrames that we want to join are passed to the merge function using the left and right argument.
The left_on='species_id' argument tells merge to use the species_id column as the join key from survey_sub (the left DataFrame).
Similarly , the right_on='species_id' argument tells merge to use the species_id column as the join key from species_df (the right DataFrame).
For inner joins, the order of the left and right arguments does not matter.
The result merged_inner DataFrame contains all of the columns from survey_sub (record_id, month, day, etc.) as well as all the columns from species_sub (species_id, genus, species, and taxa).
Left joins
What if we want to add information from species_sub to survey_df without losing any of the information from survey_sub?
In this case, we use a different type of join called a “left outer join”, or a “left join”.
Like an inner join, a left join uses join keys to combine two DataFrames.
Unlike an inner join, a left join will return all of the rows from the left DataFrame, even those rows whose join key(s) do not have values in the right DataFrame.
Rows in the left DataFrame that are missing values for the join key(s) in the right DataFrame will simply have null (i.e., NaN or None) values for those columns in the resulting joined DataFrame.
A left join is performed in pandas by calling the same merge function used for inner join, but using the how='left' argument:
merged_left = pd.merge(left=survey_sub, right=species_df, how='left', left_on='species_id', right_on='species_id')
merged_left
record_id month day year plot_id species_id sex hindfoot_length \
0 1 7 16 1977 2 NL M 32
1 2 7 16 1977 3 NL M 33
2 3 7 16 1977 2 DM F 37
3 4 7 16 1977 7 DM M 36
4 5 7 16 1977 3 DM M 35
5 6 7 16 1977 1 PF M 14
6 7 7 16 1977 2 PE F NaN
7 8 7 16 1977 1 DM M 37
8 9 7 16 1977 1 DM F 34
9 10 7 16 1977 6 PF F 20
weight genus species taxa
0 NaN Neotoma albigula Rodent
1 NaN Neotoma albigula Rodent
2 NaN Dipodomys merriami Rodent
3 NaN Dipodomys merriami Rodent
4 NaN Dipodomys merriami Rodent
5 NaN NaN NaN NaN
6 NaN Peromyscus eremicus Rodent
7 NaN Dipodomys merriami Rodent
8 NaN Dipodomys merriami Rodent
9 NaN NaN NaN NaN
The result DataFrame from a left join (merged_left) looks very much like the result DataFrame from an inner join (merged_inner) in terms of the columns it contains.
However, unlike merged_inner, merged_left contains the same number of rows as the original survey_sub DataFrame.
When we inspect merged_left, we find there are rows where the information that should have come from species_sub (i.e., species_id, genus, and taxa) is missing (they contain NaN values).
Other join types
The pandas merge function supports two other join types:
- Right (outer) join: Invoked by passing
how='right'as an argument.- Similar to a left join, except all rows from the
rightDataFrame are kept, while rows from theleftDataFrame without matching join key(s) values are discarded.
- Similar to a left join, except all rows from the
- Full (outer) join: Invoked by passing
how='outer'as an argument.- This join type returns the all pairwise combinations of rows from both DataFrames; i.e., the result DataFrame will
NaNwhere data is missing in one of the dataframes. - This join type is very rarely used.
- This join type returns the all pairwise combinations of rows from both DataFrames; i.e., the result DataFrame will
Key Points
Pandas is a powerful solution for tabular data
Many SQL operations have equivalents in Pandas
Pandas can read/write many data formats