Parallel Computing
Overview
Teaching: 40 min
Exercises: 40 minQuestions
What is parallel computing?
How can I do parallel computing?
Objectives
Understand the different types of parallelism
Experience the joy of parallelism with xargs
Understand different memory models for parallel processing
Intro
For many people parallel computing brings to mind optimization. Whilst optimization can often lead you down the path of parallel computing, the two are not the same. Parallel computing is about making use of resources while optimization is about making better use of the resources.
Types of parallelism
One of the biggest advantages of using an HPC is that you will have access to a large number of processing cores. These cores are often not that much faster than what you have on a desktop machine (they may even have a lower clock speed). A desktop machine will probably have between 4-12 CPU cores, where as the individual nodes of an HPC facility will have 24-48 cores, and of course there can be hundreds or thousands of such nodes within the facility. The key to making best use of HPC resources is thus organizing your work to make the best use of these multiple cores, and this means that you need to understand how to work in parallel.
There are many different levels of parallelism that you can work with, which we’ll explore now.
Task based parallelism
In task based parallelism we are working at the highest level of abstraction. In this model we take our workflow and break it into discrete tasks and then ask which tasks are independent of each other. These independent tasks can then be run at the same time in parallel. You may see these tasks referred to as embarrassingly parallel tasks.
Depending on how the software is implemented there are two main ways to run tasks in parallel:
- Run one task per compute node, with many nodes being use at once. This would typically mean running an array job.
- Run one task per CPU within a single compute node, with just one node being used at a time. This would be facilitated with a single job script.
We can of course run hybrid models which combine the two above extremes.
Suppose we have 1000 tasks that need to be done, and each task can be completed using a single CPU core in 6hours.
Suppose we want to run this on gadi, such that we have 107 compute nodes, each with 48 CPU cores .
We could use (1) above, but it would required 1,000 nodes x 6 hours of resources (6,000 node hours), and only use 1/48th of the available cores available on each node.
This is not a good use of resources.
We can’t use (2) above because our nodes don’t have 1000 cores in any single node.
Instead we could divide our 1,000 tasks into 1000/48 = ~ 21 jobs, and then within each job run one task per CPU (assuming it will fit within the memory constraints of the node).
This could be achieved by having a job array of 21 jobs, and each job would orchestrate the running of tasks numbered (n-1)*21 to (n-1)*21 + n, and making sure that the last job in the array stops at the 1,000th task.
This method will result in 21 nodes x 6 hours of resources, or ~48 better than the previous option.
We refer to this type of organization as job packing.
Here is a visualization of what we are doing:
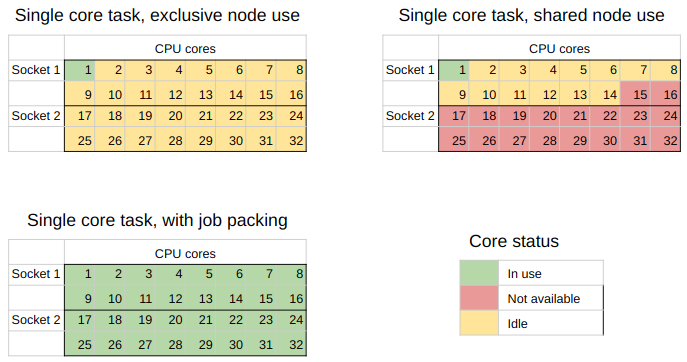 In the top left we have the worst case scenario where a single user uses a node to run a task that uses just one core of the node.
In the top right is a more realistic view of what happens when node resources can be shared.
Multiple users can run jobs on the same node, but it’s not common that the requests people make are going to add up to 100% resource usage so there will be some wastage.
In the bottom left is the goal that we are aiming for with job packing - complete use of all the CPU cores for the tasks we are running.
In the top left we have the worst case scenario where a single user uses a node to run a task that uses just one core of the node.
In the top right is a more realistic view of what happens when node resources can be shared.
Multiple users can run jobs on the same node, but it’s not common that the requests people make are going to add up to 100% resource usage so there will be some wastage.
In the bottom left is the goal that we are aiming for with job packing - complete use of all the CPU cores for the tasks we are running.
Occasionally, the limiting resource on a compute node is not the number of CPU cores but the amount of RAM available. If we can either limit or estimate the peak RAM usage of our tasks then we can adjust our job packing such that we use as much of the RAM as possible per node. For example, we might have 192GB of RAM available per node, but have a task that uses up to 10GB of RAM, and so we’d only be able to pack 19 copies of this task onto each of our compute nodes.
If we instead had 10,000,000 tasks that each took 20 second to run this would still have a total run time of approx 6,000 node hours, the same requirement as the previous example. However, PBSPro takes some time to schedule a job, allocate resources, and then clean up when the job completes. This time is not that long (maybe 30 seconds), but if your job has a relatively short run time (few minutes) then this overhead becomes a significant overhead. In such cases we can pack our jobs such that we have 10,000 jobs running within a single batch job, but only 32 running at one time. This is a second form of job packing.
In each of the above implementations the parallelism is being handled by a combination of PBSPro and our batch job scripts. We don’t need to have any knowledge of (or access to) the source code of the program that is actually doing the compute.
Job packing with xargs
The program xargs is standard on most Unix based systems and was created to “build and execute command lines from standard input”.
At its most basic level, xargs will accept input from STDIN and convert this into commands which are then executed in the shell.
xargs is able to manage the execution of these sub processes that it spawns and thus can be used to run multiple programs in parallel.
We will again simulate a hard task by doing something simple and then sleeping.
In this case we have a script called greet.sh (here) which is as follows:
greet.sh#! /usr/bin/env bash echo "$@ to you my friend!" sleep 1
If we were to have a file which consisted of greetings (greetings.txt, here), one per line, we could use xargs to run our above script with the greeting as an argument:
xargs -a greetings.txt -L 1 -exec ./greet.sh
The -L 1 instructs xargs to pass one line at a time as arguments to our -exec command, and -a indicates the input data file.
The above would eventually output the following:
Hello to you my friend!
Gday to you my friend!
Kaya to you my friend!
Kiaora to you my friend!
Aloha to you my friend!
Yassas to you my friend!
Konnichiwa to you my friend!
Bonjour to you my friend!
Hola to you my friend!
Ni Hao to you my friend!
Ciao to you my friend!
Guten Tag to you my friend!
Ola to you my friend!
Anyoung haseyo to you my friend!
Asalaam alaikum to you my friend!
Goddag to you my friend!
Shikamoo to you my friend!
Namaste to you my friend!
Merhaba to you my friend!
Shalom to you my friend!
You’ll see that the sleep 1 command means that each greeting is followed by a pause, and that we only get one greeting at a time.
The code is being executed on a single CPU core sequentially.

If we want to work with 8 tasks in parallel we can do so using the -P 8 argument to xargs:
xargs -a greetings.txt -L 1 -P 8 -exec ./greet.sh
You’ll see that we get the same output as before (maybe in a different order) but that it occurs in batches of 8, with an approximately 1 second pause between them.
What is happening now is that the waiting time is happening in parallel rather than in serial.
If we replaced the sleep 1 command with some actual work that needs to be done then we’d be making use of multiple cores in no time!
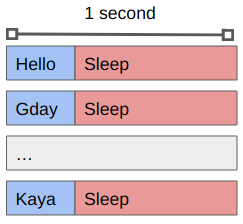
By using xargs we can create a single job file that will spawn multiple tasks (up to some maximum) that will run concurrently.
Moreover, if we have more tasks to complete than CPU cores available, xargs will wait for a task to complete before starting another.
Vectorized operations
Vectorization is the process of rewriting a loop so that instead of doing one operation per loop over N loops, your processor will do the same operation on multiple data simultaneously per loop.
Modern CPUs provide support for this via what is called single instruction multiple data (SIMD) instructions. A CPU with a 512 register can hold 16x 32bit numbers at once (or 8x 64bits), and apply the same instruction to all of them within a single clock cycle.
For example, if you wanted to add two vectors together, you could have a loop like this:
for (int i=0; i<16; i++)
C[i] = A[i] + B[i];
Which would be executed on your CPU as like this if there is no vectorization happening:
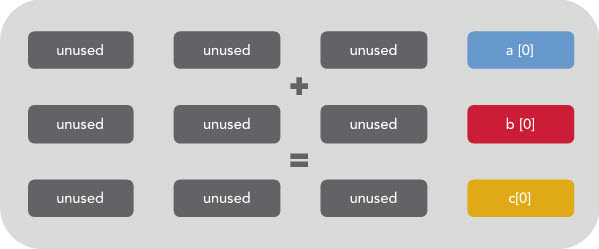
You can see that only 1/4 of the register is being used. The rest is effectively wasted. However, if vectorization is enabled then the CPU will execute the same instruction on multiple data at once like this:
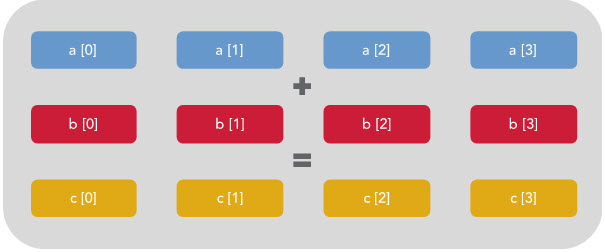
The above example is written in C rather than Python, because the C compiler is where this vectorization occurs. When compiled with the right flags, the C preprocessor will figure out which loops can be vectorized and rewrite them according to the data type and size / availability of the CPU register of the system that you are working on.
In a language like Python there is no compiler, just an interpreter, so how do we make use of vectorization?
The answer is that we take our Python code, write it in C or Fortran, and then make a python wrapper function that will package our data up and send it to that code for processing.
This is all rather fiddly work that most people don’t have time to do, so instead you should rely on libraries like numpy or scipy which are really python interfaces to fast, optimized, and often vectorized, C libraries.
Note that vectorization isn’t magic and it has some limits. The key thing to remember is that SIMD stands for SAME instruction multiple data. If your loop isn’t doing an identical operation every time then you can’t make use of vectorization. Things that break vectorization include: loop dependencies (accessing values after they have been changed in a previous iteration), flow control (if/break/continue statements within the loop), and calling functions within the loop.
In Python, the good rule of thumb is that vectorization is as simple as using numpy data structures and replacing your loops with to calls to numpy.
Vectorization with numpy
We’ll continue the above example with a python loop that computes C=A+B, where A and B are lists or arrays of integers.
In this example we’ll show how we can complete the same operation using either Python list objects or numpy.array objects.
Add two python lists
Using
ipythondo the following and observe the output:A_list=list(range(10_000)) B_list=list(range(10_000)) %timeit C_list = [ a+b for a,b in zip(A_list,B_list)]Output
Depending on the speed of your computer you’ll get something like this:
529 µs ± 21.8 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
Add two numpy arrays
Again using
ipython, do the following and observe the output:# assuming the same session as before import numpy as np A = np.array(A_list) B = np.array(B_list) %timeit C = A+BOutput
Depending on the speed of your computer you’ll get something like this:
1.18 µs ± 44.1 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)
numpy contains more than just basic math functions.
In fact many of the linear algebra operations that you would want to perform on arrays, vectors, or matrices (in the numpy.linalg module), call on powerful system level libraries such as OpenBLAS, MKL, and ATLAS.
These libraries, in turn, are multi-threaded or multi-core enabled, so in many cases you’ll also be able to make use of multiple cores, without having to explicitly deal with the multiprocessing library, just by using numpy or scipy functions.
Some particularly useful examples are the scipy.optimize and scipy.fft modules.
The main lesson here is that Python is slow but easy to code, and C is fast but hard(er) to code, but by using libraries such as numpy you can start to get the benefit of both worlds - easy to code, fast to use.
So, wherever possible, use already built libraries and avoid re-implementing things yourself.
Some potentially useful places to start are:
Domain or Data based parallelism
Consider an task that reads a data array, transforms it, and then writes it out again.
The simplest implementation of such a task can be represented as follows, where f(x) represents the transform, and we iterate over all the data in order:
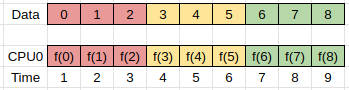 In this example we have one compute unit (CPU0) doing all the work.
In this example we have one compute unit (CPU0) doing all the work.
If multiple compute units are available (CPU0-2) then we can parallelize our work by having each compute unit perform the same set of instructions, but working on different parts of the data.
We can then have these processes running in parallel as follows, and do the same work in 1/3 the time.
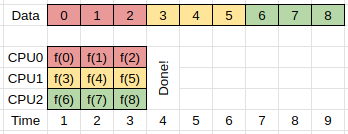
The above approach is often referred to as either domain or data based parallelism, because we are dividing our data into domains, and then working on each domain in parallel.
Here we assume that the work that needs to be done to compute f(x_i) is independent of any previous results, or rather, that the order in which the results are computed is unimportant.
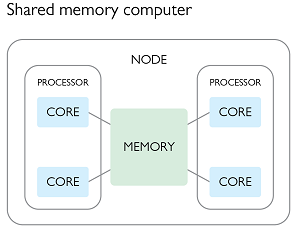
If our particular computing task falls into the above category, then we can replace our single processing design with a multiprocessing design in which all the processes that are doing the work have access to the same input/output memory locations. This form of parallelism requires that all the compute processes have access to the same memory which usually means that they all have to be on the same node of the HPC cluster that you are working on.
Another form of parallelism occurs when we have the same input data, but we want to process this data in different ways to give different outputs.
We could simply write completely different programs to perform the different calculations, but typically there is some preprocessing or setup work that needs to be done which is common between all the tasks.
Here the separate tasks are represented by the functions f(x), g(x), and h(x), and can be run simultaneously.
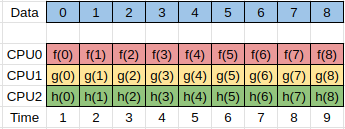
In order to be able to implement the parallelism discussed here we need to understand how to share information between different processes. This is discussed in the next section.
Memory models
We have explored some ways of doing implicit multiprocessing by taking advantage of existing tools or libraries. We are now going to look at some of the explicit ways in which we can make use of multiple CPU cores at the same time. Any time we have a program that is working across multiple cores, we will in fact be working with a collection of processes (typically 1 per core), which are communicating with each other in order to complete the task at hand. Working with multiple cores or processes thus requires that we understand how to share information between processes, and thus we will discuss the two main paradigms - shared memory, and distributed memory.
Parallel processing with shared memory
In this paradigm we create a parent program which spawns multiple child processes, each of which have access to some common shared memory. This shared memory can be used for both reading or writing. In the second figure of the previous section, we could have a parent process spawning three children for a total of three active processes, using the following plan:
- The parent process would create a shared memory location and read the input data into it, and then create an empty shared memory location for writing the output data.
- The parent process would then spawn three children and pass them a reference to the shared memory locations, as well as information about which parts of the processing they will be responsible for (e.g. start/end memory address).
- All three of the processes would then perform the same operation
f(x)on different parts of the input array, and write to different parts of the output array. - When the parent process has completed it’s work it will wait for the children to complete theirs, and then write the output data to a file.
Each of the parent and child processes would run on a separate CPU core, and have their own memory allocation in addition to the shared memory.
The beauty of the above example is that the individual processes do not need to communicate or synchronize with each other in order to complete their work. Most importantly, they are never trying to read or write from the same memory address as each other. The only constraint is that all of the processes need to be able to directly reach the memory (RAM) in question, and this typically means that they all have to be running on the same node of an HPC. The scaling of your task is thus limited by the number of cores (and memory) available on a single node of the HPC facility.
Suppose that the function to be performed was to compute a running average of the input data over some window. In this case each of the processes would need to read data from overlapping regions of the input array (at the start/end of their allocation). Since the input data is not changing, having multiple concurrent reads is not an issue so each process can continue to act in isolation.
Suppose that the function we are performing is to build a histogram of the input data. The output data are then a set of bins initiated to zero, and the “function” is to read the bin, increment the value by one, and then write the result back to the output. Each of the processes can still read the input data without interfering with each other but now need to ensure that updating the output array doesn’t cause conflicts. For example, if two processes try to increment the same bin at the same time, then the’ll both read the same value (e.g. 0), increment it (to 1), and then write the result. One of these processes will write first and the other will write second, but the second one will overwrite the first. What we need in this case is a way to indicate that the read/increment/write part of the code can only be done by one process at a time. In this case we can use what is called a lock on the memory location. A process would lock a memory address, do the required read/increment/write, and then unlock the memory address. The library which provides the shared memory will track of all the locks, and if a process asks to use some memory space which is locked the process will be forced to wait until the lock is released before doing so. To avoid creating/releasing locks thousands of times, it would instead be useful to have each of the processes create their own version of the output data to work on locally, and then do the update once per bin when they are finished.
The OpenMP library is the most widely used for providing shared memory access to C and Fortran programs. Other languages (such as python) which provide shared memory libraries are either built on OpenMP or at least use the same programming paradigm and will therefore use similar terminology, and have similar limitations to OpenMP.
In Python there are two ways to achieve parallel computing: multiprocessing, and MPI4py.
Here is an implementation of our sky_sim.py code using multiprocessing.
sky_sim_mp.py#! /usr/bin/env python # Demonstrate that we can simulate a catalogue of stars on the sky # Determine Andromeda location in ra/dec degrees import math import numpy as np import multiprocessing import sys NSRC = 1_000_000 def get_radec(): # from wikipedia ra = '00:42:44.3' dec = '41:16:09' d, m, s = dec.split(':') dec = int(d)+int(m)/60+float(s)/3600 h, m, s = ra.split(':') ra = 15*(int(h)+int(m)/60+float(s)/3600) ra = ra/math.cos(dec*math.pi/180) return ra,dec def make_stars(args): """ """ #unpack the arguments ra, dec, nsrc = args # create an empy array for our results radec = np.empty((2,nsrc)) # make nsrc stars within 1 degree of ra/dec radec[0,:] = np.random.uniform(ra-1, ra+1, size=nsrc) radec[1,:] = np.random.uniform(dec-1, dec+1, size=nsrc) # return our results return radec def make_stars_parallel(ra, dec, nsrc=NSRC, cores=None): # By default use all available cores if cores is None: cores = multiprocessing.cpu_count() # 20 jobs each doing 1/20th of the sources group_size = nsrc//20 args = [(ra, dec, group_size) for _ in range(20)] # start a new process for each task, hopefully to reduce residual # memory use ctx = multiprocessing.get_context() pool = ctx.Pool(processes=cores, maxtasksperchild=1) try: # call make_posisions(a) for each a in args results = pool.map(make_stars, args, chunksize=1) except KeyboardInterrupt: # stop all the processes if the user calls the kbd interrupt print("Caught kbd interrupt") pool.close() sys.exit(1) else: # join the pool means wait until there are results pool.close() pool.join() # crete an empty array to hold our results radec = np.empty((2,nsrc),dtype=np.float64) # iterate over the results (a list of whatever was returned from make_stars) for i,r in enumerate(results): # store the returned results in the right place in our array start = i*group_size end = start + group_size radec[:,start:end] = r return radec if __name__ == "__main__": ra,dec = get_radec() pos = make_stars_parallel(ra, dec, NSRC, 2) # now write these to a csv file for use by my other program with open('catalog.csv', 'w') as f: print("id,ra,dec", file=f) np.savetxt(f, np.column_stack((np.arange(NSRC), pos[0,:].T, pos[1,:].T)),fmt='%07d, %12f, > %12f')
Make a branch
Let’s make a new branch for each of our parallel implementations.
For multiprocessing lets make a branch called
multiprocessing:git branch multiprocessing
There are two main things that we need to do differently in this version of the code compared to our original implementation.
Firstly note that the code is largely unchanged, except for the introduction of a new function called make_stars_parallel, and that we have changed the call signature of the original function to just accept args instead of ra, dec, nsrc.
def make_stars(args):
#unpack the arguments
ra, dec, nsrc = args
# was
def make_stars(ra, dec, nsrc=NSRC):
We’ll come back to why we changed this in a minute.
The new function (make_stars_parallel) that we create is what I call the driving or wrapper function, and it is the one that handles all the multiprocessing.
The new function has the same call signature as the old function, but adds a new parameter called cores which has a default value.
This means that the new function can act as a drop-in replacement for the old one.
Within the new function we need to do the following:
- determine how many processes we are going to run
- ideally this is less than or equal to the number of physical CPU cores available
- figure out what work needs to be done
- divide the work into groups
- the number of groups is usually close to an integer multiple of the number of processes
- set up a pool of workers (child processes)
- send work teach of the workers
- wait for the work to complete
- collect the results from each worker
- return the results
Lets now run through each part:
1 determine the number of processes to use
We let the user specify a number of cores using the cores argument.
If they don’t specify a number (eg it’s value is None) then we ask the multiprocessing library to count the number of threads (physical CPUs + virtual CPUs) the CPU has available.
NB: this will sometimes be 2x the number of physical cores due to hyperthreadding or simultaneous multithreading, but we don’t mind if that happens.
# By default use all available cores
if cores is None:
cores = multiprocessing.cpu_count()
2 figure out what work needs to be done
For the sky_sim program we are simulating a large number of points around a single location.
The central location is ra,dec and the number of points is nsrc=NSRC (supply by the user).
3 divide the work into groups
We can divide the task among n processes by having each of them compute nsrc/n points and then collecting all the generated points at the end.
In our example we choose to divide the work into 20 batches, but we could also choose n or some integer multiple of n.
For each batch of work we need to set up the argument list that we’ll be sending to the make_stars function.
# 20 jobs each doing 1/20th of the sources
group_size = nsrc//20
args = [(ra, dec, group_size) for _ in range(20)]
Note that in our list of args, each element is a tuple.
4 set up a pool of workers
To manage all the processes that we are going to create and use, we need a context manager which we get from the multiprocessing module.
From this manager we can then create a Pool of workers.
We specify how many processes we want to use and how many tasks each child process will execute.
For our simple case we want only 1 task per child, this means that when a task is complete the python instance will shut down and new one will be started within that process.
This helps to avoid any issues related to us not cleaning up after ourselves when we are done (memory leaks, residual state, etc).
# start a new process for each task, hopefully to reduce residual
# memory use
ctx = multiprocessing.get_context()
pool = ctx.Pool(processes=cores, maxtasksperchild=1)
We could also have asked for a Queue instead of a Pool, the difference being that in a Queue, work is returned in the order in which you put it into the queue, where as in a pool the work is returned in the order of completion.
A pool will be more effective at keeping the child processes engaged, but a queue will let you have your results in the order you sent them to be processed.
For our task we don’t care about ordering so we use a pool.
Note that we could still use a pool if we cared about the ordering, we’d just have to tell each worker what it’s position was in the queue, and then have that position returned to us as part of the results.
From here we could reconstruct the intended ordering of the results.
5 send work teach of the workers
We now have work to do and workers to complete it so we join them together.
We could submit jobs to the pool one at a time using pool.
We use the map method to apply the fucntion make_stars to each of the items in the iterable args.
results = pool.map(make_stars, args, chunksize=1)
The reason that we had to rewrite the call sign for make_stars is because pool.map will only take one of the items from args at a time, so we bundled them into tuples.
The child processes are now being created and will start working.
6 wait for the work to complete
Because the various processes are all doing their own thing, and this parent process will continue executing code in the mean time, we have to tell the parent process to wait until all the others are complete before we try to retrieve the results.
We do this by calling pool.join(), but we must first call pool.close() to indicate that no more work is going to be submitted to the pool.
# join the pool means wait until there are results
pool.close()
pool.join()
7 collect the results from each worker
Each of or workers have executed the make_stars function, which returns a tuple of (ras,decs) and we now want to collect them all together into a single larger array.
We could create empty python lists using ras=[] and append our results to these lists.
However, appending to a list in python gets slower as the list gets longer.
If we already know how long the list will be in the first place we should just create a list with that length.
Since the data coming back from the child processes are numpy arrays, we’ll create a new numpy array with the right size and then squish all the data into that.
We could use np.zeros(shape, dtype) to create an empty array, but since we are going to fill/assign every value in that array, we can get a very small performance boost by just asking for memory without setting it to zero.
We do this with np.empty(shape, dtype)
# crete an empty array to hold our results
radec = np.empty((2,nsrc),dtype=np.float64)
And now we go about stuffing the results into the corresponding location in the new array.
# iterate over the results (a list of whatever was returned from make_stars)
for i,r in enumerate(results):
# store the returned results in the right place in our array
start = i*group_size
end = start + group_size
radec[:,start:end] = r
8 return the results
This is easy :D
return radec
what about all the other code?
In our description of 1-8 we missed a bunch of code.
What does it do?
Extra bits
There are a few good practices that we should obey when doing our multiprocessing, and this is where the extra code comes in.
Firstly, we should plan for when things go bad.
If we were running our program, and the user presses
How are the processes communicating with each other?
In our example above, the different processes don’t actually share any memory. They share information between each other, but keep their own separate memory spaces. The information is shared by inter process communication. In this simple example there are only two points at which the processes need to communicate:
- when the child processes begin they need to be told which code to execute and on which data
- when the child processes complete their work, they have to send the results back
Python does this communication using serialization, which means that the data are converted from python objects into strings, the strings are communicated between processes, and then the strings are turned back into objects.
Python uses pickle to do this serialization.
The things that we need to know here is that:
- serialization is slow (-er than just sending memory addresses)
- a serialized object takes more memory than the underlying object
- passing data via strings is inefficient
In our example, the amount of time taken for the child processes to create the random data was much smaller than the time taken to pass the data back to the parent process. If we were to spread the work over many cores, it might actually take more time to complete the work thanks to this message passing overhead. In reality, however, we would have a more complicated simulation process so that the message passing time was a small fraction of the total compute time.
There is an alternative way for us to manage the results that are being returned that will avoid almost all of the serialization work, and that is to use a properly shared memory model.
Thankfully the multiprocessing module also provides us with some shared memory functions.
Shared memory in python
The shared memory that is implemented by the multiprocessing library relies on an underlying C library to do all the work.
The consequence of this is that we are limited in the types of memory that can be shared.
We can use:
multiprocessing.Valuemultiprocessing.Arraymultiprocessing.shared_memory.SharedMemory
For the Value and Array data types we have to use CTypes data types which you refer to using special type codes.
For the SharedMemory data type you are sharing a block of memory, and it’s up to you to figure out how to deal with it.
The advantage of these shared memory objects is that when you change their value in one process, all processes will see the updated value. (But beware of race conditions!).
A very nice feature is that numpy arrays can be instructed to use a SharedMemory objects memory as the storage location for the data.
This means that you can create a numpy array that is shared between multiple processes.
Let’s look at how we can do that in another example.
sky_sim with (proper) shared memory
sky_sim_sharemem.py#! /usr/bin/env python # Demonstrate that we can simulate a catalogue of stars on the sky # Determine Andromeda location in ra/dec degrees import math import numpy as np import multiprocessing from multiprocessing.shared_memory import SharedMemory import uuid import sys NSRC = 1_000_000 mem_id = None def init(mem): global mem_id mem_id = mem return def get_radec(): # from wikipedia ra = '00:42:44.3' dec = '41:16:09' d, m, s = dec.split(':') dec = int(d)+int(m)/60+float(s)/3600 h, m, s = ra.split(':') ra = 15*(int(h)+int(m)/60+float(s)/3600) ra = ra/math.cos(dec*math.pi/180) return ra,dec def make_stars(args): """ """ ra, dec, shape, nsrc, job_id = args # Find the shared memory and create a numpy array interface shmem = SharedMemory(name=f'radec_{mem_id}', create=False) radec = np.ndarray(shape, buffer=shmem.buf, dtype=np.float64) # make nsrc stars within 1 degree of ra/dec ras = np.random.uniform(ra-1, ra+1, size=nsrc) decs = np.random.uniform(dec-1, dec+1, size=nsrc) start = job_id * nsrc end = start + nsrc radec[0, start:end] = ras radec[1, start:end] = decs return def make_stars_sharemem(ra, dec, nsrc=NSRC, cores=None): # By default use all available cores if cores is None: cores = multiprocessing.cpu_count() # 20 jobs each doing 1/20th of the sources args = [(ra, dec, (2, nsrc), nsrc//20, i) for i in range(20)] exit = False try: # set up the shared memory global mem_id mem_id = str(uuid.uuid4()) nbytes = 2 * nsrc * np.float64(1).nbytes radec = SharedMemory(name=f'radec_{mem_id}', create=True, > size=nbytes) # creating a new process will start a new python interpreter # on linux the new process is created using fork, which > copies the memory # However on win/mac the new process is created using spawn, > which does # not copy the memory. We therefore have to initialize the > new process # and tell it what the value of mem_id is. method = 'spawn' if sys.platform.startswith('linux'): method = 'fork' # start a new process for each task, hopefully to reduce > residual # memory use ctx = multiprocessing.get_context(method) pool = ctx.Pool(processes=cores, maxtasksperchild=1, initializer=init, initargs=(mem_id,) # ^-pass mem_id to the function 'init' when > creating a new process ) try: pool.map_async(make_stars, args, chunksize=1).get> (timeout=10_000) except KeyboardInterrupt: print("Caught kbd interrupt") pool.close() exit = True else: pool.close() pool.join() # make sure to .copy() or the data will dissappear when > you unlink the shared memory local_radec = np.ndarray((2, nsrc), buffer=radec.buf, dtype=np.float64).copy() finally: radec.close() radec.unlink() if exit: sys.exit(1) return local_radec if __name__ == "__main__": ra, dec = get_radec() pos = make_stars_sharemem(ra, dec, NSRC, 2) # now write these to a csv file for use by my other program with open('catalog.csv', 'w') as f: print("id,ra,dec", file=f) np.savetxt(f, np.column_stack((np.arange(NSRC), pos[0, :].T, > pos[1, :].T)),fmt='%07d, %12f, %12f')
Make another branch
For this version of the code lets make a branch called
mp-sharemem:git branch mp-sharememNote that we are now branching a branch.
mp-sharememis branched frommultiprocessingwhich is branched frommain!
A quick summary of what is different this time (compared to our serial version):
- we define a global variable (
mem_id) which will indicate the shared memory location - we have modified
make_starsto have an altered call signature (as before)make_starsno longer returns any data, but instead writes it directly to shared memory
- we have a wrapper function
make_stars_sharememthat will handle the creation of shared memory, creating child process, and then dishing out work.
The overview of what we are doing is slightly different from before. Below is the process with the main changes in bold:
- determine how many processes we are going to run
- ideally this is less than or equal to the number of physical CPU cores available
- figure out what work needs to be done
- divide the work into groups
- the number of groups is usually close to an integer multiple of the number of processes
- create some shared memory
- set up a pool of workers (child processes)
- send work teach of the workers
- wait for the work to complete
- copy data from shared memory back to local memory and de-allocate the shared memory
- return the results
Let’s look at the steps that have changed.
4 create some shared memory
In the parent process we first create a random name for our memory obejct.
For this I like to use the universally unique identifier uuid package.
# set up the shared memory
global mem_id
mem_id = str(uuid.uuid4())
Next we have to figure how how much memory we will need.
If you have only worked in python, then this will be an unfamiliar concept, because python normally just expands memory to suit your needs.
However, the SharedMemory is closely bound to the underlying C implementation and changing the size of the memory once created is not allowed.
Since the size of memory is determined also by the data type we have to think a little more carefully about that also.
In this case we are going to eventually want an np.array that has shape (2,NSRC) and has data type np.float64.
nbytes = 2 * nsrc * np.float64(1).nbytes
radec = SharedMemory(name=f'radec_{mem_id}', create=True, size=nbytes)
In the above we create the shared memory with the create=True option.
Within the child nodes, which are going to run make_stars we’ll have to do a similar call, but this time with create=False
# Find the shared memory and create a numpy array interface
shmem = SharedMemory(name=f'radec_{mem_id}', create=False)
Again in the make_posistions function, we want to treat the shared memory as if it were a numpy array so we do the following:
radec = np.ndarray(shape, buffer=shmem.buf, dtype=np.float64)
since we need to know the shape of the numpy array in the make_stars function, we have to pass that as one of the arguments to the function.
One final modification we make to the make_stars function, is to save the results into this shared memory.
In order to not step on the toes of any other process, we have to know where abouts this data should be written.
In our example we use the job_id (process number) to figure out where abouts to write the data.
start = job_id * nsrc
end = start + nsrc
radec[0, start:end] = ras
radec[1, start:end] = decs
We could have done the above copy without using numpy arrays, but it involves a lot of python loops and there are many opportunities to get the addresses wrong. Being able to slice a numpy array makes this easier.
5 set up a pool of workers
Now that we are using shared memory we have a piece of information that is going to be passed to each of the child workers, which is the name (address) of the shared memory.
There are two ways to do this, either add a new argument to the make_stars function (easy!), or use an initializer function (harder but demonstrated for completeness).
In unix based operating systems (including older MacOS) new processes are created using fork which means that they are an exact copy of the parent process, and have a copy of the parent memory.
Unfortunately, Windows and new MacOS, don’t support fork and instead use spawn to create new processes.
When a new process is spawned, the memory of the parent is not copied, so we need to do work to copy across the required information.
Linux also supports spawn, but fork is much faster so we would like to use it if possible.
We create an initializer function called init that looks like:
def init(mem):
global mem_id
mem_id = mem
return
We decide on the method for creating new processes:
method = 'spawn'
if sys.platform.startswith('linux'):
method = 'fork'
And then we create a new context manager and pool.
As we create the pool, we tell it that the initializer function is init and that the arguments are the variable mem_id:
ctx = multiprocessing.get_context(method)
pool = ctx.Pool(processes=cores, maxtasksperchild=1,
initializer=init, initargs=(mem_id,)
)
8 copy data from shared memory back to local memory and de-allocate the shared memory
Once all the child processes complete, we have all the information that we need in shared memory.
To copyt this sharedmemory into a local numpy array we use a similar trick as before, but we then append .copy().
# make sure to .copy() or the data will dissappear when you unlink the shared memory
local_radec = np.ndarray((2, nsrc), buffer=radec.buf,
dtype=np.float64).copy()
Once we have copied the data we de-allocate the shared memory:
radec.close()
radec.unlink()
If we don’t do the above then the memory will not (neccessarily) be deallocated. Python usually complains that there was some still allocated shared memory around when your program exits, and will try to release it, but I don’t 100% trust it. (And it’s good practice to clear up after yourself!)
what about all the other code?
In our description of 1-8 we missed a bunch of code.
What does it do?
more extra bits
This time we have two try clauses.
The first one is to make sure that the additional processes are cleaned up when we quit the program early.
The second try clause is an interesting one that you might not have seen before:
try:
...
finally:
radec.close()
radec.unlink()
if exit:
sys.exit(1)
The finally clause is executed after everything in the try/except/else has been sorted out, even if there were exceptions being thrown.
This means that even if something blows up in the try clause, we will eventually come back to the finally clause and do all the cleanup.
Here the cleanup is to close the shared memory object (stops further access), and then unlink it (de-allocates the memory).
In order to ensure that this finally clause is hit, we move the sys.exit(1) from the inner try/except to here.
How are we going?
Did all that sink in?
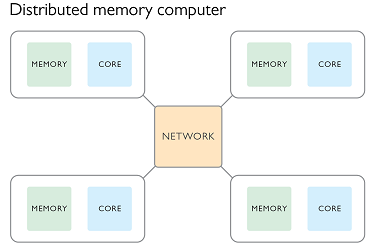
Parallel processing with distributed memory
In this paradigm we create a number of processes all at once and pass to them some meta-data such as the total number of processes, and their process number. Typically the process numbered zero will be considered the parent process and the others as children. In this paradigm each process has it’s own memory and there is no shared memory space. If we wanted to repeat the simple computing example used previously we would use the following plan:
- All processes use their process number to figure out which part of the input data they will be working on.
- Each process reads only the part of the input data that they require.
- Each process computes
f(x)on the input data and stores the output locally. - Each child process sends their output data to the parent process.
- The parent process creates a new memory allocation large enough to store all the output data, and copies it’s own output into this memory.
- The parent process then receives the output data from each child process and copies it to the output data array.
- The parent process writes the output to a file.
- All processes are now complete and terminate.
Each of the processes would run on a different CPU core and have their own memory space. This makes it possible for different processes to be run on different nodes of an HPC facility, with the message passing being done via the network.
It is still possible to have each process running on the same node. A message passing interface (MPI) has been developed and implemented in many open source libraries. As the name suggests the focus is not on sharing memory, but in passing information between processes. These processes can be on the same node or different nodes of an HPC.
The key to understanding how an MPI based code works is that all the processes are started simultaneously and then connect to a communication hub (usually called COMM_WORLD), they then execute the code. During code execution a processes can send/recieve messages from any/all of the other nodes. A special type of message that is often used is called a barrier, which causes each process to block (wait) until all process are at the same point in the code. This is often required because, despite starting at the same time, the different processes can quickly get out of sync, especially if they need to perform I/O, communicate over a network, or simply have different data that they are working on.
Unlike with multiprocessing, the number of processes being used is determined outside of our code so we always have to ask questions like “how many processes are there total?” and “what is my process number?”.
The process number is refered to as it’s rank and the only rank that is guaranteed to exist is 0, so this is often used as the special/parent rank.
Continuing our sky_sim example we can use MPI to acheive our simulation task.
sky_sim_mpi.py#! /usr/bin/env python # Demonstrate that we can simulate a catalogue of stars on the sky # Determine Andromeda location in ra/dec degrees import math import numpy as np from mpi4py import MPI import glob NSRC = 1_000_000 comm = MPI.COMM_WORLD # rank of current process rank = comm.Get_rank() # total number of processes that are running size = comm.Get_size() def get_radec(): # from wikipedia ra = '00:42:44.3' dec = '41:16:09' d, m, s = dec.split(':') dec = int(d)+int(m)/60+float(s)/3600 h, m, s = ra.split(':') ra = 15*(int(h)+int(m)/60+float(s)/3600) ra = ra/math.cos(dec*math.pi/180) return ra,dec def make_stars(ra, dec, nsrc, outfile): """ """ # make nsrc stars within 1 degree of ra/dec ras = np.random.uniform(ra-1, ra+1, size=nsrc) decs = np.random.uniform(dec-1, dec+1, size=nsrc) # return our results with open('{0}_part{1:03d}'.format(outfile, rank), 'w') as f: if rank == 0: print("id,ra,dec", file=f) np.savetxt(f, np.column_stack((np.arange(nsrc), ras, decs)),fmt='%07d, %12f, %12f') return if __name__ == "__main__": ra,dec = get_radec() outfile = "catalog_mpi.csv" group_size = NSRC // size make_stars(ra, dec, group_size, outfile) # synchronize before moving on comm.Barrier() # Select one process to collate all the files if rank == 0: files = sorted(glob.glob("{0}_part*".format(outfile))) with open(outfile,'w') as wfile: for rfile in files: for l in open(rfile).readlines(): print(l.strip(),file=wfile) # os.remove(rfile)
Make yet another branch
Since our MPI implementation isn’t related to the multiprocessing ones, we’ll make a new branch but this time branch off main.
git checkout main git branch mpi
To run the above code we use a syntx similar to xargs:
mpirun -n 4 python3 sky_sim_mpi.py
mpirun not working?
You’ll nee to have an mpi library installed. On ubuntu you can use mpich which is installed as:
sudo apt install mpichIf your local machine doesn’t have mpi you can install it, but sometimes it can be rather tricky. Instead you might just log into gadi and run your work there.
module load openmpi
Whilst we can use our COMM_WORLD to pass messages between processes, it’s a very ineffective way to pass large quantities of data (see previous chat about serialization). One way to get around this issue is to have each process dump their work products in a shared directory and then have another program join all the work together. Sometimes this “other” program is just the rank 0 process. This is the approach that we’ll be taking today.
With MPI we will need to do the following in our script:
- figure out how many processes are running
- figure out what work needs to be done in the current process
- do the work
- write the output to a file
- communicate that the work is done
- ONE process (rank=0) needs to collect all the work into a single file
1 figure out how many processes are running
To do this we connect to the COMM_WORLD and ask for the rank of this process:
comm = MPI.COMM_WORLD
# rank of current process
rank = comm.Get_rank()
# total number of processes that are running
size = comm.Get_size()
2 figure out what work needs to be done in the current process
Again we are going to assume that each process does 1/n of the work.
This time we don’t need a complicated wrapper script, we just compute the amount of work done in our if __name__ clause:
group_size = NSRC // size
3/4 do the work + write the output to a file
Since each process will be writing it’s own output file we pass a filename to our make_stars function along with the usual ra, dec, nsrc.
Aditionally, we can’t use the same file name for all processes so we have to modify the filename for each process:
def make_stars(ra, dec, nsrc, outfile):
...
# return our results
with open('{0}_part{1:03d}'.format(outfile, rank), 'w') as f:
if rank == 0:
print("id,ra,dec", file=f)
np.savetxt(f, np.column_stack((np.arange(nsrc), ras, decs)),fmt='%07d, %12f, %12f')
return
Note that we stuck the .csv header into the first file which is being written by the rank 0 process.
5 communicate that the work is done
In each process, when the work is done and we have returned from the make_stars function we call:
make_stars(ra, dec, group_size, outfile)
# synchronize before moving on
comm.Barrier()
This will cause all the processes to wait until they are all at the same line of code.
6 collect all the work into a single file
This only needs to be done by one process so we choose the rank 0 process. Within this process we read all the files that were written by the other processes, and then write them out in order.
# Select one process to collate all the files
if rank == 0:
files = sorted(glob.glob("{0}_part*".format(outfile)))
with open(outfile,'w') as wfile:
for rfile in files:
for l in open(rfile).readlines():
print(l.strip(),file=wfile)
# os.remove(rfile)
We are also going to tidy up all the partial files here (last line).
What about the … ?
There were no extra bits this time.
Notes
There are two inefficiencies that we have in the above code:
- the rank 0 process wrote it’s data to a file and then read it in again
- we had to wait for all processes to finish before we started writing the combined file
- If we did some point to point communication we could tell rank 0 to start reading the rank 1 file as soon as it was ready
MPI can sometimes be a much simpler approach than shared memory, even when you are working on multiple cores of the same node. Sometimes MPI is an absolute brain destroyer when you are trying to debug it, because all your debug messages overlap or appear out of order.
At the end of the day you should chose a solution that will work for your use case which includes one that you can implement in a reasonable amount of time. (Your time is more valuable than computer time!)
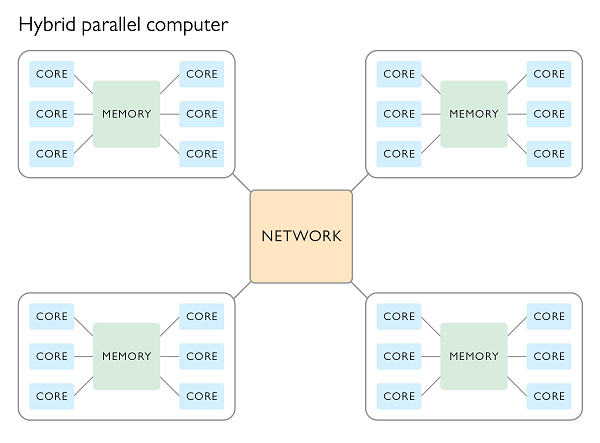
Hybrid parallel processing
It is possible to access the combined CPU and RAM of multiple nodes all at once by making use of a hybrid processing scheme. In such a scheme a program will use MPI to dispatch a bunch of primary processes, one per node, which in turn control multiple worker processes within each node which share memory using OpenMP.
Summary
There are many levels of parallelism that can be leveraged for faster throughput.
The type of parallelism used will depend on the details of the job at hand or the amount of time that you are able and willing to invest.
Starting with the easy parts first (eg job arrays and job packing with xargs) and then moving to shared memory or MPI jobs until you reach a desired level of performance is recommended.
Push your work to github
For each of the branches that you have created:
git checkout [branch]git pushThen go onto github and view the different branches.
Key Points
Parallelism is not about optimization per se
Understanding your workflow is key to planning your parallel processing
Start with what is easy, make things harder only when necessary
If using python leverage numpy data structures and functions