How to Automate the Boring Bits
Overview
Teaching: 45 min
Exercises: 45 minQuestions
How can automation improve my current workflow?
How can I automate my current work?
How can I ensure that my workflow is reproducible on different computers?
What workflow management tools are available?
Objectives
Learn to streamline the routine tasks in your research - from downloading remote data to managing reproducible computational environments - so you can focus on discovery.
Overview
Why automate?
Automation helps streamline repetitive tasks, reduce errors, and save time, allowing you to focus on analysis and discovery. By incorporating automation techniques into your research workflows, you can improve the efficiency, consistency, and reproducibility of your work, ultimately leading to more reliable and impactful scientific discoveries.
Common repetitive tasks in research:
Research often involves a variety of repetitive tasks that can benefit from automation, including:
- Data retrieval: Regularly downloading large datasets from remote servers or online repositories.
- Data preprocessing: Cleaning, filtering, and formatting raw data to prepare it for analysis.
- Data analysis: Running the same analysis scripts on multiple datasets or subsets of data.
- Report generation: Creating standardized reports or visualizations from analysis results.
- Environment setup: Configuring computational environments with specific software dependencies for different projects.
How automation can help:
- Efficiency: Automating repetitive tasks can save significant time and effort, allowing you to focus on more complex and creative aspects of your work.
- Consistency: Automated workflows ensure that tasks are performed the same way every time, reducing the risk of human error and increasing the reliability of results.
- Reproducibility: By automating the setup of computational environments and the execution of analysis scripts, you can easily reproduce your work and share it with others.
- Scalability: Automation makes it easier to scale up analyses to larger datasets or more complex workflows without a proportional increase in manual effort.
- Documentation: Automated workflows often include built-in documentation and logging, making it easier to track what was done and troubleshoot any issues that arise.
Tools and techniques for automation:
- Scripting languages: Use languages like Python or Bash to write scripts that automate repetitive tasks.
- Workflow management tools: Tools like Make and Nextflow can help manage complex workflows with multiple dependencies and parallel tasks.
- Version control: Use Git to track changes to scripts and workflows, ensuring that you can revert to previous versions if needed.
- Continuous integration: Set up continuous integration (CI) pipelines to automatically run tests and analyses whenever changes are made to your code or data.
Leveraging Large Language Models (LLMs) for Automation:
Large Language Models (LLMs) like ChatGPT and (GitHub) CoPilot can assist in automating various aspects of research by providing intelligent suggestions, generating code snippets, and even automating documentation tasks. As a bonus, many of these are free for academic use, or are included as part of your organisations enterprise software agreements. Here are some ways LLMs can be utilized:
- Code Generation: LLMs can generate code snippets for common tasks, reducing the time spent writing boilerplate code.
- Data Analysis: LLMs can assist in writing complex data analysis scripts by providing suggestions and examples.
- Documentation: Automatically generate documentation for code, workflows, and research findings, ensuring consistency and saving time.
- Testing: LLMs can suggest tests for your code, reducing the time writing boilerplate code.
Verify LLM outputs
A very important note about the use of LLMs (or any assistive tools) is that their output is not guaranteed to be: error free, up-to-date, appropriate, or logically sound. You are ultimately responsible for your research and software and so you should carefully validate the information that you get from these tools. Think of these tools as an untrusted colleague or random stack-overflow contributor with low reputation - you need to think critically about what is being presented to you.
Today’s focus
In this workshop we are going to start with a commonly disliked, overlooked, and time consuming aspect of research - data cleaning - and learn some techniques to automate the process. The scenario that are going to work on is one that I had to struggle with during my PhD - preparing catalogues of radio sources for analysis. At the time, tools such as AstroPy were just being developed, and were certainly not as widely used as today. Not only that, but the data that I was using wasn’t avaialble in consistent formats - different research groups provided data via different online services which had very different data formats. Today the world is a little nicer, as many people have realised the benefits of FAIR data not only to people who want to use the data, but also those who want to be recognised for publishing it.
The task that we will focus on today is the following:
- Obtain a catalogue of data data for a given radio survey
- Clean and filter the data
- Convert the data into a standard format
We will firstly look at how we would do all of this manually (because thats how you do everything the first time), and then learn some tools that will help us to automate the process.
A workflow for humans
- Download the AT20G dataset from this repo (a subset of the entire catalogue from ViZieR)
- Separate the meta data from the table data (starts with
#) - Remove the second two lines of the table header (units and
----) - Count the number of rows in the data, record for later use
- Correct the data types of the columns (currently most are strings, thanks to missing data)
- Remove all rows with no 5/8GHz measurements (columns S8 and S5)
- Keep only rows with 12 <= RA <= 18 (hours)
- Count the number of rows rejected/remaining, record for later use
- Delete all columns from Ep -> Sp
- Save the filtered file for analysis work as
AT20G_clean.csv
What tools can we use?
Excel or similar is a good enough tool, widely available, that you can use to inspect and edit your data.
If a new version of that catalogue comes out (same format, adjusted values or more rows), how long does it take to redo this?
If you need to add extra filtering steps, how much work do you need to redo?
When you write up your methods section and need to describe this filtering process, where have you recorded it? How do you know how many rows passed each criteria?
You want to change your analysis so that you can see the distribution of sources on sky before/after the filtering is done. Do you have a dataset that would allow this?
Let’s now take our workflow and make it a workflow for computers.
A workflow for computers
For each of the steps above we need to identify a tool.
Overall we have three main things we are doing with our data:
- Downloading data
- Separating lines of a file based on content
- Filtering rows / columns of a table
While there are many different options available, and we could potentially use one tool to do everything, I find it a lot easier to choose the right tool for the right job. This can mean you need to learn a few different tools, but you don’t need to become expert in each tool.
The tools that we are going to use today are:
wgetorcurlfor downloading filesgrepfor selecting lines of a file based on contentpython3/pandasfor manipulating our table
Downloading data from remote sources
wget and curl are two programs that are usually available on most systems.
They both perform the same task - connect to a url and saves the content to a file.
If you hit a url which is a website, you’ll end up with a .html file.
If you connect to a file then you’ll get the file itself.
Download our AT20g.tsv file using either of the following options (depending on what is available on your system).
# Download a single file
wget https://raw.githubusercontent.com/ADACS-Australia/2025-ASA-ECR-WorkshopSeries/refs/heads/gh-pages/data/Workshop1/AT20G.tsv
You should see some output like this, and the AT20G.tsv file in your current directory.
--2025-04-17 11:49:35-- https://raw.githubusercontent.com/ADACS-Australia/2025-ASA-ECR-WorkshopSeries/refs/heads/gh-pages/data/Workshop1/AT20G.tsv
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.110.133, 185.199.109.133, 185.199.108.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.110.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 550370 (537K) [text/plain]
Saving to: ‘AT20G.tsv’
AT20G.tsv 100%[===================>] 537.47K --.-KB/s in 0.005s
2025-04-17 11:49:36 (106 MB/s) - ‘AT20G.tsv’ saved [550370/550370]
And if we are using curl, the syntax is almost the same:
# Download a single file
curl -O https://raw.githubusercontent.com/ADACS-Australia/2025-ASA-ECR-WorkshopSeries/refs/heads/gh-pages/data/Workshop1/AT20G.tsv
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 537k 100 537k 0 0 20.5M 0 --:--:-- --:--:-- --:--:-- 20.1M
Again the file will end up in the current directory.
Now that we have figured out how to download the file automatically, we should copy the commands that we used for reference later on.
Make a workflow file
Make a file called
workflow.txtwhich we’ll use to create our automated workflow. Add thewget/curlcommand that you used above into this file.
Separating data from meta-data
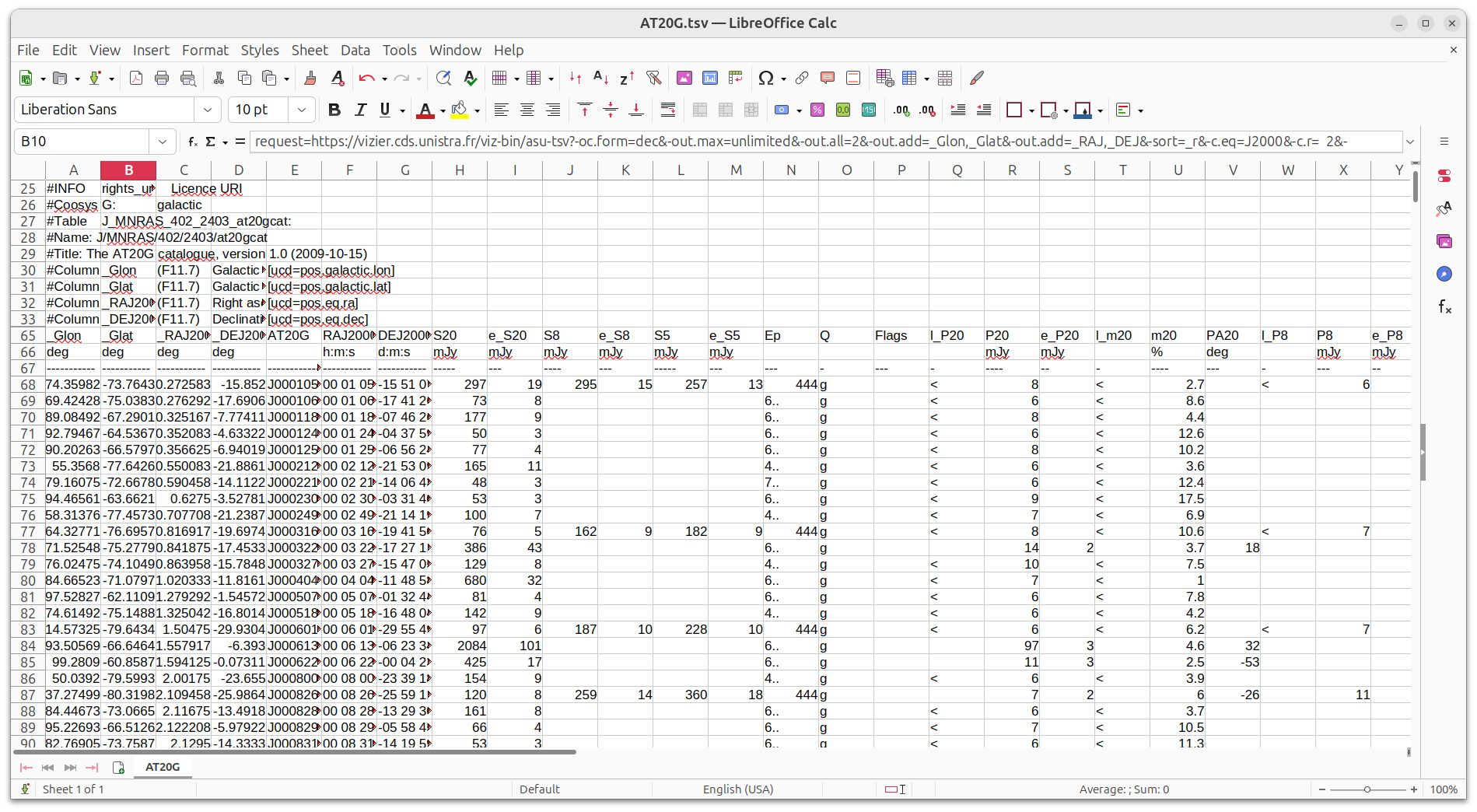
We should now have a file called AT20G.tsv in our working directory, which looks like the following:
AT20G.tsv
# # VizieR Astronomical Server vizier.cds.unistra.fr # Date: 2025-04-10T06:17:11 [V7.4.6] # In case of problem, please report to: cds-question@unistra.fr # # #Coosys J2000: eq_FK5 J2000 #INFO service_protocol=ASU IVOID of the protocol through which the data was retrieved #INFO request_date=2025-04-10T06:17:11 Query execution date #INFO request=https://vizier.cds.unistra.fr/viz-bin/asu-tsv?-oc.form=dec&-out.max=unlimited&-out.all=2&-out.add=_Glon,_Glat&-out.add=_RAJ,_DEJ&-sort=_r&-c.eq=J2000&-c.r= 2&-c.u=arcmin&-c.geom=r&-source=J/MNRAS/402/> 2403/at20gcat&-order=I&-out=AT20G&-out=RAJ2000&-out=DEJ2000&DEJ2000=>-30 & <0&-out=S20&-out=e_S20&-out=S8&-out=e_S8&-out=S5&-out=e_S5&-out=Ep&-out.all=2&-out=Q&-out=Flags&-out=l_P20&-out=P20&-out=e_P20&-out=l_m20&-out=m20&-out=PA20&-out=l_P8&-out=P8&-out=e_P8&-out=l_m8&-out=m8&-out=PA8&-out=l_P5&-out=P5&-out=e_P5&-out=l_m5&-out=m5&-out=PA5&-out=Sp&Sp=Sp&-out.all=2& Full request URL (POST) ... #Column PA5 (I3) [-90/90]? Position angle at 5GHz [ucd=pos.posAng] #Column Sp (A2) SPECFIND of radio surveys with this source (Vollmer et al., Cat. VIII/85) [ucd=meta.ref.url] _Glon _Glat _RAJ2000 _DEJ2000 AT20G RAJ2000 DEJ2000 S20 e_S20 S8 e_S8 S5 e_S5 Ep Q Flags l_P20 P20 e_P20 l_m20 m20 PA20 l _P8 P8 e_P8 l_m8 m8 PA8 l_P5 P5 e_P5 l_m5 m5 PA5 Sp deg deg deg deg "h:m:s" "d:m:s" mJy mJy mJy mJy mJy mJy mJy mJy % deg mJy m Jy % deg mJy mJy % deg ----------- ----------- ----------- ----------- -------------- ----------- ----------- ----- --- ---- --- ----- --- --- - --- - - --- -- - ---- --- - --- -- - ----- --- - ---- - - ----- --- -- 074.3598240 -73.7643280 000.2725833 -15.8520000 J000105-155107 00 01 05.42 -15 51 07.2 297 19 295 15 257 13 444 g < 8 < 2.7 < 6 < 2.0 < 6 < 2.3 Sp 069.4242750 -75.0383103 000.2762917 -17.6906111 J000106-174126 00 01 06.31 -17 41 26.2 73 8 6.. g < ... 093.5244966 -61.6186061 359.3548750 -1.8704444 J235725-015213 23 57 25.17 -01 52 13.6 124 6 6.. p < 10 < 8.0 Sp 081.4390180 -69.8275875 359.3803750 -11.4274444 J235731-112538 23 57 31.29 -11 25 38.8 804 38 6.. p 12 2 1.5 -86 Sp 087.0646275 -67.0857923 359.5407500 -8.0010278 J235809-080003 23 58 09.78 -08 00 03.7 67 3 6.. p < 6 < 9.0 Sp 083.6389933 -69.0391655 359.5458333 -10.3355556 J235811-102008 23 58 11.00 -10 20 08.0 1243 59 6.. p < 12 < 1.0 Sp 021.2857573 -78.1419382 359.5700417 -28.8926944 J235816-285333 23 58 16.81 -28 53 33.7 147 10 219 12 178 9 444 p < 6 < 4.3 < 9 < 4.1 < 6 < 3.4 Sp 058.1557506 -76.5292565 359.8314583 -20.7988333 J235919-204755 23 59 19.55 -20 47 55.8 100 4 222 7 257 8 444 p e < 7 < 10.4 < 13 < 12.9 < 7 < 4.7 Sp 089.5191701 -66.1071830 359.8830000 -6.6621944 J235931-063943 23 59 31.92 -06 39 43.9 113 6 6.. g < 8 < 7.2 Sp 095.7351629 -60.6265524 359.9041667 -0.5206389 J235937-003114 23 59 37.00 -00 31 14.3 272 14 7.. g 6 1 2.3 0 Sp
The structure of the file is as follows:
- a lot of metadata (rows starting with #)
- header information for the table columns (three rows)
- data for the table with tabs separating each entry
In order to separate the metadata rows we’ll use a program called grep which is great for selecting/deselecting lines of a file based on a string pattern match.
Select all the lines that start with a
#grep -e '^#' AT20G.tsvHere we are telling
grepto print out all the rows of the given file that have a#as the first character on the line (^represents the start of the line). Type this into your terminal and review the output.
Save the header to a separate file
We can direct the output of
grepinto a file using the redirection operator>. For example:grep -e '^#' > AT20G_header.txtDo this now and check that the new file exists and has all the meta data in it.
If we want to make another file which has the meta-data stripped from it, we can instruct grep to return all non matchin rows using the -v operator.
grep -v -e '^#' AT20G.tsv
We could pipe these results to an intermediate file for further processing, however grep allows us to chain many patterns together.
Thus we can have a single command like the following for our filtering:
grep -v -e '^#' -e '^-' -e '^deg' -e '^$' AT20G.tsv
Where we use the following:
-vshow NON matching rows-eadd expression for matching^#match lines starting with # (the metadata)^deglines starting with deg (the second line in the header)^-match lines starting with - (the thrid line in the header)^$empty lines (^and$represent the first/last char in a line, so^$is a shortcut for empty lines)
Note: the ordering of the match patterns doesn’t affect the content of the output.
Use less to page output
The previous
grepcommand will produce pages of output, and the key results that we are looking for are on the first page only. We can page the output by piping the results to a program calledless:<command> | less(press Q to close the program)
I recommend you do this with the above
grepcommand to see that the header is correct.
Update your workflow file
Use the
grepfunctions that we worked with above, to add two new lines to your workflow file:
- Save the meta-data to a file called
AT02G_header.txt- Save the table data to a file called
AT20G_table.tsv
my workflow file
# Download the data wget https://raw.githubusercontent.com/ADACS-Australia/2025-ASA-ECR-WorkshopSeries/refs/heads/gh-pages/data/Workshop1/AT20G.tsv # Save the meta-data grep -e '^#' AT20G.tsv > AT20G_header.txt # Save just the table with a 1 line header grep -v -e '^#' -e '^-' -e '^deg' -e '^$' AT20G.tsv > AT20G_table.tsvNote that I’ve added some comments (starting with #) to this file so that I can recall what the different commands are supposed to be doing.
Cleaning the table data
The command line tools we used so far are great for simple options, but tring to filter a table based on string matching is a very hard task.
Therefore we will reach into our toolbox for another tool, this time the pandas module in python.
For this next section to work you’ll need to have python3 installed with both the pandas and numpy modules.
numpy is a dependency for pandas so you can just install pandas and you’ll have everything you need.
To check if you have pandas installed you can run the following:
python3 -c 'import pandas'
If you get no output then everything is fine, if you get an error message then you’ll need to have pandas installed using pip install pandas or conda install pandas, depending on how your python environment is set up.
We’ll work from the command line for now so lets open up python and import the modules that we’ll be using:
import pandas as pd
import numpy as np
load our table with
pandasUse the pd.read_csv function to load the table and then view a summary of the table.
pandasrefers to tables as a “data frame” sodfis the most commonly used as the name for this variable.df = pd.read_csv('AT20G_table.tsv', delimiter='\t') df_Glon _Glat _RAJ2000 _DEJ2000 AT20G RAJ2000 DEJ2000 S20 e_S20 S8 ... l_m8 m8 PA8 l_P5 P5 e_P5 l_m5 m5 PA5 Sp 0 74.359824 -73.764328 0.272583 -15.852000 J000105-155107 00 01 05.42 -15 51 07.2 297 19 295 ... < 2.0 < 6 < 2.3 Sp 1 69.424275 -75.038310 0.276292 -17.690611 J000106-174126 00 01 06.31 -17 41 26.2 73 8 ... Sp 2 89.084918 -67.290098 0.325167 -7.774111 J000118-074626 00 01 18.04 -07 46 26.8 177 9 ... Sp 3 92.794672 -64.536666 0.352083 -4.633222 J000124-043759 00 01 24.50 -04 37 59.6 50 3 ... Sp 4 90.202631 -66.579685 0.356625 -6.940194 J000125-065624 00 01 25.59 -06 56 24.7 77 4 ... Sp ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... .. 2831 83.638993 -69.039165 359.545833 -10.335556 J235811-102008 23 58 11.00 -10 20 08.0 1243 59 ... Sp 2832 21.285757 -78.141938 359.570042 -28.892694 J235816-285333 23 58 16.81 -28 53 33.7 147 10 219 ... < 4.1 < 6 < 3.4 Sp 2833 58.155751 -76.529257 359.831458 -20.798833 J235919-204755 23 59 19.55 -20 47 55.8 100 4 222 ... < 12.9 < 7 < 4.7 Sp 2834 89.519170 -66.107183 359.883000 -6.662194 J235931-063943 23 59 31.92 -06 39 43.9 113 6 ... Sp 2835 95.735163 -60.626552 359.904167 -0.520639 J235937-003114 23 59 37.00 -00 31 14.3 272 14 ... Sp [2836 rows x 35 columns]The first (un-named) column is the index, which is not part of our original file, but something that
pandashas added. We’ll just ignore the index for now.
Our table is a little bit broken at the moment, because the input data file has some missing data. If we view the data types of the different columns we’ll see the following:
>>> df.dtypes
_Glon float64
_Glat float64
_RAJ2000 float64
_DEJ2000 float64
AT20G object
RAJ2000 object
DEJ2000 object
S20 object
e_S20 int64
S8 object
e_S8 object
S5 object
e_S5 object
Ep object
...
Sp object
dtype: object
The first four columns are all of type float64 which means numeric data, however most of the remainder are of type object which means they are strings.
We don’t care about most of the columns, but the columns S20, S8, and S5 (and the corresponding e_ columns) are going to be used in our later analysis and should be numeric, but are currently strings.
If we look at the data in these columns (see previous challenge), you can see that some of the S8 and S5 data are missing.
Unfortunately this data set represents missing data as a bunch of spaces rather than using an empty cell.
eg. the table looks like \t\s\s\s\t instead of just \t\t for the missing data, and so pandas interprets the entire column as being strings.
The first thing that we want to do is fix this annoyance.
Instead of a string of spaces we want the blank cells to be represented with np.nan which signals to pandas that we don’t have data for that cell.
We’ll use df.replace to find all the cells that are empty or one or more spaces and replace them with np.nan. We can use some of our grep knowledge to help with this filtering.
Recall that we used ^$ to represent an empty line in our file earlier, well that pattern is what we call a regular expression (or regex).
I like to think of regex as a meta-tool that is useful for a lot of different applications.
For example we could write r'^\s*$' to represent the pattern of “a string of zero or more spaces, and nothing else”.
Replace blank strings with
np.nanUse
df.replace(r<pattern>, <value>, regex=True)to replace all the strings of spaces withnp.nanYou should see output that looks like the following if you get it right:
_Glon _Glat _RAJ2000 _DEJ2000 AT20G RAJ2000 DEJ2000 S20 e_S20 S8 ... l_m8 m8 PA8 l_P5 P5 e_P5 l_m5 m5 PA5 Sp 0 74.359824 -73.764328 0.272583 -15.852000 J000105-155107 00 01 05.42 -15 51 07.2 297 19 295 ... < 2.0 NaN < 6 NaN < 2.3 NaN Sp 1 69.424275 -75.038310 0.276292 -17.690611 J000106-174126 00 01 06.31 -17 41 26.2 73 8 NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN Sp 2 89.084918 -67.290098 0.325167 -7.774111 J000118-074626 00 01 18.04 -07 46 26.8 177 9 NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN Sp 3 92.794672 -64.536666 0.352083 -4.633222 J000124-043759 00 01 24.50 -04 37 59.6 50 3 NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN Sp 4 90.202631 -66.579685 0.356625 -6.940194 J000125-065624 00 01 25.59 -06 56 24.7 77 4 NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN Sp ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... .. 2831 83.638993 -69.039165 359.545833 -10.335556 J235811-102008 23 58 11.00 -10 20 08.0 1243 59 NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN Sp 2832 21.285757 -78.141938 359.570042 -28.892694 J235816-285333 23 58 16.81 -28 53 33.7 147 10 219 ... < 4.1 NaN < 6 NaN < 3.4 NaN Sp 2833 58.155751 -76.529257 359.831458 -20.798833 J235919-204755 23 59 19.55 -20 47 55.8 100 4 222 ... < 12.9 NaN < 7 NaN < 4.7 NaN Sp 2834 89.519170 -66.107183 359.883000 -6.662194 J235931-063943 23 59 31.92 -06 39 43.9 113 6 NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN Sp 2835 95.735163 -60.626552 359.904167 -0.520639 J235937-003114 23 59 37.00 -00 31 14.3 272 14 NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN Sp [2836 rows x 35 columns]Once you get the desired output, save that as a new table
df_fix.Solution
df_fix = df.replace(r'^\s*$', np.nan, regex=True)
Unfortunately updating the content of the columns doesn’t change the data types (check with df_fix.dtypes), so we have to do that explicitly.
We can do this using the df[<colname>].astype(float) function on each of the columns.
The following loop will replace each of the columns that we care about with a numercial version of that column.
for colname in ['S20', 'e_S20', 'S8', 'e_S8', 'S5', 'e_S5']:
df_fix[colname] = df_fix[colname].astype(float)
Once you run this you should see the same output when showing the table (just type df_fix), but the column data types should change (as see with df_fix.dtypes)
We now have a table with all the null values correctly represented and the correct data types for the columns that we are interested in.
Summarise data using
.describe()
pandasgives us a nice method for giving a quick summary of our (numeric) data. Use the.describe()method on thedf_fixtable to see how many of the columns have null values in them, as well as the distribution of data.Solution
>>> df_fix.describe() _Glon _Glat _RAJ2000 _DEJ2000 S20 e_S20 S8 e_S8 S5 e_S5 P20 m20 count 2836.000000 2836.000000 2836.000000 2836.000000 2832.000000 2836.000000 959.000000 959.000000 961.000000 961.000000 2836.000000 2836.000000 mean 181.477329 -13.517460 168.684580 -15.047620 224.745410 12.538434 330.374348 16.875912 358.907388 17.491155 11.029619 8.861248 std 106.677853 41.256409 107.098922 8.649608 617.360857 33.853303 507.352544 25.850951 498.699643 23.818633 24.258990 6.484771 min 0.013309 -88.693811 0.272583 -29.998833 40.000000 2.000000 5.000000 1.000000 3.000000 1.000000 6.000000 1.000000 25% 63.723809 -51.176564 76.593437 -22.367993 71.000000 4.000000 107.000000 6.000000 106.000000 6.000000 6.000000 4.400000 50% 214.304398 -14.184359 156.615646 -15.372653 105.000000 6.000000 177.000000 9.000000 194.000000 10.000000 7.000000 7.400000 75% 253.293749 24.806519 262.480625 -7.594056 184.000000 10.000000 335.000000 16.000000 399.000000 20.000000 11.000000 11.500000 max 359.937922 61.995122 359.904167 -0.002056 20024.000000 996.000000 5735.000000 299.000000 4424.000000 221.000000 1074.000000 76.500000
If we want to save the number of null values in a given column we can ask the question more directly:
df_fix['S8'].isnull().sum()
Filtering the table data
Now that we have cleaned the data, we are in a position to start slicing and dicing our data. Pandas makes this easy as it lets us use an array of True/False to index the table, which sets us up for a powerful idiom called masking. The typical masking idiom looks like this
mask = df.where(
We can do things like:
- select not null data using
mask = ~df_fix['S5'].isnull() - Select only data based on an equation
mask = df_fix['_RAJ2000'] > 12*15) - Combine masks using binary operators (
& | ~)mask = df_fix['_RAJ2000'] > 12*15) & (df_fix['_RAJ2000']<18*15) - Drop columns via selection
df_fix = df_fix[ <list of column names to keep>]
Filter the table
Use the masking idiom examples to select only data such that:
- The value of
S8andS5is not null.- The value of
_RAJ2000is between 12 and 18 hours (it’s stored as degrees)- Keep only the following columns:
['_Glon', '_Glat', '_RAJ2000', '_DEJ2000', 'AT20G', 'RAJ2000', 'DEJ2000', 'S20', 'e_S20', 'S8', 'e_S8', 'S5', 'e_S5']Solution
mask = ~(df_fix['S5'].isnull() | df_fix['S8'].isnull()) mask = mask & ((df_fix['_RAJ2000'] > 12*15) & (df_fix['_RAJ2000']<18*15)) df_fix = df_fix[mask] df_fix = df_fix[['_Glon', '_Glat', '_RAJ2000', '_DEJ2000', 'AT20G', 'RAJ2000', 'DEJ2000', 'S20', 'e_S20', 'S8', 'e_S8', 'S5', 'e_S5']] df_fix.describe()>>> df_fix.describe() _Glon _Glat _RAJ2000 _DEJ2000 S20 e_S20 S8 e_S8 S5 e_S5 count 181.000000 181.000000 181.000000 181.000000 181.000000 181.000000 181.000000 181.000000 181.000000 181.000000 mean 320.735786 35.494619 209.284729 -22.701123 241.176796 15.149171 312.220994 16.419890 329.922652 16.872928 std 17.900302 7.068314 17.003349 4.374341 372.634050 23.995714 410.828669 20.872375 402.125871 19.497019 min 286.646694 7.449553 180.888750 -29.998833 42.000000 3.000000 35.000000 3.000000 8.000000 1.000000 25% 305.204633 30.882807 194.727167 -26.568000 87.000000 6.000000 107.000000 6.000000 100.000000 6.000000 50% 321.261922 36.221876 208.626500 -22.838833 137.000000 8.000000 189.000000 10.000000 196.000000 10.000000 75% 335.642502 41.058205 222.760375 -18.432472 214.000000 13.000000 334.000000 16.000000 398.000000 20.000000 max 353.729587 46.592231 255.290625 -15.051222 3449.000000 226.000000 3647.000000 190.000000 3173.000000 158.000000
Now that we have a cleand and filtered table we can write out a new file using the .to_csv function of our data frame.
Remember that index that pandas added?
We’ll be removing that as we write the output file.
We should also change the format to be .csv (comma separated values) instead of .tsv (tab separated values), since different systems render tabs as a different number of characters (and they look like spaces).
This will make the file easier for humans to read and spot mistakes, while not making it any harder for computers to read the files.
Save our data
Use
df_fix.to_csv('AT20G_final.csv', index=False)to save our cleaned and filtered table to a new file.
Now we have figured out all the steps needed to do our cleaning and filtering with Python, but we don’t want to have to use the command line like that all the time, because it’s gross, takes a lot of time, and is easy to make mistakes.
Create a python script for our data cleaning
Trace back through all the commands that you executed in order to get to the final output. Choose the commands that worked, and copy them into a new file called
clean_AT20G.py. Commands likedf_fixordf.describe()that only print data can be mostly skipped over. Bonus marks if you add some short comments (start with#) to describe what your script is doing.
clean_AT20G.pyimport pandas as pd import numpy as np # Read the table df = pd.read_csv('AT20G_table.tsv', delimiter='\t') # replace all the spaces with nulls and change the column types df_fix = df.replace(r'^\s*$', np.nan, regex=True) for colname in ['S20', 'e_S20', 'S8', 'e_S8', 'S5', 'e_S5']: df_fix[colname] = df_fix[colname].astype(float) # filter out all the rows with null S8/S5 and keep only those in a given RA range mask = ~(df_fix['S5'].isnull() | df_fix['S8'].isnull()) mask = mask & ((df_fix['_RAJ2000'] > 12*15) & (df_fix['_RAJ2000']<18*15)) df_fix = df_fix[mask] # drop the columns that we don't need df_fix = df_fix[['_Glon', '_Glat', '_RAJ2000', '_DEJ2000', 'AT20G', 'RAJ2000', 'DEJ2000', 'S20', 'e_S20', 'S8', 'e_S8', 'S5', 'e_S5']] # save to a file df_fix.to_csv('AT20G_final.csv', index=False)You can check that your script works by deleting the output file and then running
python clean_AT20G.py, it should remake the file.
Update your
workflow.txtUpdate
workflow.txtto include the python based cleaning / filtering step.my workflow file
# Download the data wget https://raw.githubusercontent.com/ADACS-Australia/2025-ASA-ECR-WorkshopSeries/refs/heads/gh-pages/data/Workshop1/AT20G.tsv # Save the meta-data grep -e '^#' AT20G.tsv> AT20G_header.txt # Save just the table with a 1 line header grep -v -e '^#' -e '^-' -e '^deg' -e '^$' AT20G.tsv > AT20G_table.tsv # Clean and filter with python python clean_AT20G.py
The workflow.txt file that you have now gives you a way to reproduce all the steps of the workflow simply by copy pasting the commands into a bash terminal.
Even easier than this we could rename the file to be workflow.sh and it would be a valid bash script that we could run with bash workflow.sh!
Pretty easy right?
Create executable workflow file (optional)
If you add a
#! /usr/bin/env bashto the first line of your workflow file, then linux will interpret all of the following lines as bash code and execute them as such. Additionally, if you change the file permissions to be ‘executable’ usingchmod ugo+xthen you’ll be able to run your script from the command line by calling it by name (./workflow.sh).
We have now captured all the work of our human workflow into something that a computer can follow. Now if we want to redo any or all of this work we can just run the script. If a collaborator or supervisor asks “what if …”, you can test that out by modifying the workflow and re-running, if you don’t like the result, you can just change back and re-run, all without having to spend a alot of time. If we want someone else to be able to replicate our work we can send them the workflow file, and they should be able to run the same process on the same data. Well, this would be nice, but there is one little wrinkle that we have to deal with - the other person may not have the same software on their computer. This is a very common problem, but there are many tools available to help us get around it. We will explore one of these tools in the next section.
Managing your workflow’s environment
You want to make sure that each time you run the workflow you get the same results.
This includes when you upgrade your computer, or when you run on a different computer.
To do this, we need to not only control the workflow instructions but also the software that is being used.
For tools like wget and grep, there are not so many versions that we need to bother with, people either have the tool or they don’t.
However, for tools like python, there are many version of python itself (3.8-3.13 are commonly used), and then you have all the separate libraries which have their own versions.
In our workflow that we developed above we used two python libraries: pandas and numpy.
We’ll now look at ways that we can indicate to people which libraries are required to run our workflow, and how to make sure they are automatically installed.
Virtual environments
On your computer you will typically have many different projects that you are working on, and many different software packages installed.
One solution is that you can try and maintain a universal set of software that will support each of your projects, however this can become difficult or impossible to achieve as different programs will have different requirements.
Not only do you have your own projects that you have to worry about but many operating systems keep a version of python and associated libraries that are integral for the smooth running of the OS.
If you updated or break this system version of python you are in a world of pain.
A virtual environment is essential for managing dependencies and ensuring reproducibility in your workflows. It allows you to create an isolated Python environment for your project, separate from the global Python installation. This isolation ensures that:
-
Dependency Management: You can install specific versions of libraries required for your project without affecting other projects or the system-wide Python installation.
-
Reproducibility: By isolating dependencies, you can ensure that your workflow runs consistently across different systems, even if the global environment changes.
-
Conflict Avoidance: Virtual environments prevent version conflicts between libraries used in different projects. For example, one project might require
pandas==1.3.0, while another needspandas==1.5.0. -
Ease of Collaboration: By sharing a
requirements.txtfile, collaborators can recreate the exact environment needed to run your workflow, avoiding “it works on my machine” issues. -
Safe Experimentation: You can safely test new libraries or versions without risking the stability of your global Python environment.
Using tools like venv or conda, you can easily create and manage virtual environments tailored to your project’s needs. This practice is a cornerstone of reproducible and maintainable research workflows.
Making a python virtual environment
There are a few different tools that you can use to make a python virtual environment, and they all essentially do the same thing, just with different amounts of user-friendly-ness.
The option that takes the least amount of installation is the python library venv.
The main steps for making the environment are:
- Create a new environment:
python3 -m venv env - Activate the environment:
- On macOS and Linux:
source env/bin/activate - On Windows:
env\Scripts\activate
- On macOS and Linux:
- Install required packages:
pip install numpy pandas - Deactivate the environment:
deactivate
When you create a new environment you’ll see a new directory with the same name.
This directory contains all the information needed to run and maintain that environment.
Common names are just env (or .env), but you can use anything you like.
When you activate a new environment you’ll typically see that your command prompt changes to include (env) at the start, so you know which environment is active.
Any libraries that you install when the environment is active will only be installed into that environment.
create a new virtual environment for your project
Create a new enviroment called
.envin your project directory, activate the project, and install both pandas and numpy.
To make the environment reproducible on other computers we don’t need to copy the entire .env directory structure, instead we just need a list of which libraries were installed and their versions.
Once we have this we can make a file requirements.txt which lists (one per line) the libraries and versions.
Create a
requirements.txtfile for your projectWe installed
pandasandnumpyfor our scripts to run, so list these in a new file calledrequirements.txt.To determine the versions of these libraries we can either remember what we were told when they were installed (too hard), or we can ask
pipto tell us usingpip freeze.Find the versions of
pandasandnumpythat were installed and then add this to therequirements.txtfile.my
requirements.txtfilenumpy==2.2.4 pandas==2.2.3
Note
When you ran
pip freezeabove, you probably saw a LONG list of libraries that were installed. This is becausepandasandnumpyeach have other libraries that they rely on, and those have dependencies as well. We don’t need to exactly match every library version, just the top level libraries that we used.
To install libraries into an environment using this file, run:
pip install -r requirements.txt
In our directory we now have a few files and directories:
./
../
AT20G.tsv
AT20G_header.txt
AT20G_table.tsv
AT20G_final.csv
clean_AT20G.py
env/
requirements.txt
workflow.sh
To make it easier to remember how all the files interact and what the purpose of this proejct is we should introduce some project structure.
Project structure
Structuring your files and directories properly is crucial for maintaining a clean, organized, and manageable Python project. A well-structured project makes it easier to navigate, understand, and collaborate with others. Here are some key points to consider:
-
Root Directory: The root directory should contain essential files like
README.md,setup.py, and a license file. These files provide important information about the project, installation instructions, and licensing details. -
Source Code Directory: Create a dedicated directory (e.g.,
srcor the project name) for your source code. This directory should contain all the Python modules and packages related to your project. -
Tests Directory: Include a separate directory (e.g.,
tests) for your test cases. Organizing tests in a dedicated directory helps ensure that they are easily accessible and maintainable. -
Configuration Files: Store configuration files (e.g.,
config.yaml,.env) in a dedicated directory (e.g.,config). This keeps configuration settings separate from the source code and makes it easier to manage different environments. -
Data Directory: If your project involves data processing, create a directory (e.g.,
data) to store raw and processed data files. This helps keep data organized and prevents cluttering the source code directory. -
Documentation Directory: Maintain a directory (e.g.,
docs) for project documentation. This can include user guides, API references, and other relevant documentation. -
Virtual Environment: Use a virtual environment to manage dependencies. Create a directory (e.g.,
.env) for the virtual environment to ensure that dependencies are isolated and do not interfere with other projects.
Example project structure:
my_project/
├── env/
├── config/
│ └── config.yaml
├── data/
│ ├── raw/
│ └── processed/
├── docs/
│ └── index.md
├── src/
│ └── main.py
├── tests/
│ └── test_main.py
├── README.md
├── requirements.txt
└── setup.py
Now when we return to our project after some time away (or share it with a collaborator) we can get an idea of what is going on by first looking at the README.md file, and then poking around the various directories. Not all of the directores and files listed above are essntial for every project, however it is a good idea to consider all 7 items above and choose what you think is relevant and helpful.
Update our project structure
- Create a
datadirectory with sub-directories of raw/intermediate/final- Create a
srcdirectory for storing code- Create a
README.mdfile with a short description of the project.- Move all the data files into the relevant directories.
- Update your
clean_AT20G.pyscript to read/write using the new locations.- Update your
workflow.shfile so that it will work with the new directory structure.Note that you’ll have to update the paths to the various files now that you have moved them!
Updating
wgetto save the file in a different locationUse
wget -O path/to/file.txt urlto tellwgetto save the file in a given location.
Further workflow automation
A single script will suffice for simple and short running workflows. If the workflow breaks part way, you can make an adjustment and then restart from the begninning without it taking up much of your time. However, as your workflow gets more complicated and takes longer to run, you might not want to “restart and run all” every time you make a change. For example, you update the cleaning and filtering script, but you don’t want to download the data again.
In the remainder of this workshop we’ll explore the next level of automation which is an orchestration tool called make.
Make
Make is a build automation tool that helps manage and execute workflows by defining a series of tasks and their dependencies. It is particularly useful for scientific research because:
- Simplicity: Makefiles are straightforward to write and understand, making it easy to define workflows.
- Dependency Management: Make ensures that tasks are executed in the correct order based on their dependencies, which is crucial for reproducible research.
- Efficiency: Make only re-executes tasks that have changed, saving time and computational resources.
- Portability: Makefiles can be shared and executed on different systems, ensuring consistency across environments.
Creating a Simple Workflow with Make**
# Define targets and dependencies
all: clean_data.csv
clean_data.csv: raw_data.csv
python preprocess.py raw_data.csv clean_data.csv
# Define a clean target to remove generated files
clean:
rm -f clean_data.csv
With the above script we have a number of entry points:
- the default entry point “all”, which will kick off our workflow.
- a dummy entrypoint “clean” which removes all the intermediate/final data products to give us a “clean” slate.
- a specific file that we wish to create.
make was dessigned for managing the compilation of code in languages like c/c++/Fortran, and assumes that each stage in the workflow involves the creation of a file (target), which may or may not rely on the existance of other files (prerequisites).
A makefile describes each task as:
<target>: <prerequisite1> <prerequesite2> ...
instructions to create target file
Note: The indentation is a single tab character.
Our workflow so far is:
# Download the data
wget -O data/raw/AT20G.tsv https://raw.githubusercontent.com/ADACS-Australia/2025-ASA-ECR-WorkshopSeries/refs/heads/gh-pages/data/Workshop1/AT20G.tsv
# Save the meta-data
grep -e '^#' data/raw/AT20G.tsv > data/final/AT20G_header.txt
# Save just the table with a 1 line header
grep -v -e '^#' -e '^-' -e '^deg' -e '^$' data/raw/AT20G.tsv > data/processing/AT20G_table.tsv
# Clean and filter with python
python src/clean_AT20G.py
We can break down the above process by tracking the files that arecreated as follows:
<no file>-> AT20G.tsv- AT20G.tsv -> AT20G_header.txt
- AT20G.tsv -> AT20G_table.tsv
- AT20G_table.tsv -> AT20G_final.csv
Create a makefile for our workflow
Use the demonstrated syntax and our breakdown of the workflow to write a makefile.
Note that the characters
#and$mean something to make so we have to escape them using\#and$$respectively.
makefile# List all the outputs that we want all: data/final/AT20G_final.csv data/final/AT20G_header.txt data/raw/AT20G.tsv: wget -O data/raw/AT20G.tsv https://raw.githubusercontent.com/ADACS-Australia/2025-ASA-ECR-WorkshopSeries/refs/heads/gh-pages/data/Workshop1/AT20G.tsv data/final/AT20G_header.txt: data/raw/AT20G.tsv grep -e '^\#' data/raw/AT20G.tsv > data/final/AT20G_header.txt data/processing/AT20G_table.tsv: data/raw/AT20G.tsv grep -v -e '^\#' -e '^-' -e '^deg' -e '^$$' data/raw/AT20G.tsv > data/processing/AT20G_table.tsv # Note that I put the script as a dependency so that this step is redone if the script is updated :D data/final/AT20G_final.csv: data/processing/AT20G_table.tsv src/clean_AT20G.py python src/clean_AT20G.py # Delete all the data files clean: rm data/*/AT20G*
Our makefile requires that we have loaded the right python environment, so we should note this in our README.md file so that users don’t get hit with errors they don’t understand.
You’ll notice in the above that we often have a pattern of using the input/ouptut filenames as part of the dependencies. Make gives us some shortcuts to reduce the amount of repetition:
$<is the first prerequisite$^is the all the prerequesites$@is the target of the rule- many more listed here
This all sounds nice and fun, however these shortcuts quickly loose all meaning when you step away from the project, and make quickly becomes a write-only language.
Additionally, the inability to visualise the execution path of a given makefile, means that make is good only for small or simple projects.
This shouldn’t be seen as a criticism of make since we are essentially hijacking it for uses that it was not intended.
make was designed to make compiling code easier, not for orchestrating complex scientific workflows!
For larger or more complex projects we should therefore use something that was designed as a workflow manager.
Nextflow
Nextflow was designed and built as a workflow orchestration tool, with a focus on portability and reproducibility. Nextflow focuses on channels of information, which can be either files or variables, making it easier to pass information between different processing steps. Some of the very nice features of Nextflow that come out of the box include:
- Caching of results, and the ability to resume a previous run
- Integration with any architecture that you use (eg. HPC, cloud, desktop)
- Management of a range of containerisation tools (eg. Docker, Singularity)
- Detailed, automatic, and easily understood reporting of every run
- Monitoring of resources on a per-task basis
Nextflow is a fantastic tool, but not something we can fully learn in the remaining time for this workshop. However, we’ll have a quick overview of how it works, and you can refer to this ADACS workshop for a self-paced workshop.
Example Nextflow workflow
process preprocess {
input:
path raw_data
output:
path "clean_data.csv"
script:
"""
python preprocess.py $raw_data clean_data.csv
"""
}
workflow {
raw_data = file("raw_data.csv")
clean_data = preprocess(raw_data)
}
Session 2 topics
Your workflow will eventually fail. Actually, it will most likely fail before it ever succeeds, this is just how development works. A key time-saving practice is to be able to know where and why your workflow has failed so that you can get to fixing it. Therefore, tracking the progress of the workflow is essential, as is stopping the workflow as soon as something goes wrong. You should therefore spend some time implementing logging and error handling for your workflow / scripts.
Logging
Logging is a crucial aspect of any workflow for several reasons:
- Debugging: Logs provide detailed information about the application’s execution, which helps developers identify and fix bugs more efficiently.
- Monitoring: Logs allow you to monitor the application’s behavior in real-time, ensuring that it is running as expected and helping to detect any anomalies or issues early.
- Auditing: Logs can serve as an audit trail, recording important events and actions taken by users or the system. This is particularly useful if you want to know how your program ran, and what inputs were used, without having to run it again.
- Performance Analysis: By analyzing logs, you can identify performance bottlenecks and optimize the application’s performance.
Example: Logging with Python’s logging Module
import logging
# Configure logging
logging.basicConfig(filename='workflow.log', level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')
def log_example():
logging.info("This is an info message")
logging.warning("This is a warning message")
logging.error("This is an error message")
# Example usage
log_example()
Error handling
Use error handling to manage and recover from failures. This can include retrying failed tasks, logging errors for later analysis, and implementing checkpointing to save intermediate results. Effective error handling ensures that the workflow can continue or be easily restarted in case of issues.
In python there are a few ways to exit and signal failure.
You can raise an exception that is never caught (eg raise Exception(Things are bad)), and this will result in your program showing a traceback of where the exception was raised, and the program returning an exit code of not zero (failure).
Another option that you have is to send a message to the user (using print, or logging), and then quit the program early using os.exit(1).
You can use python to wrap or call other programs from the command line using subprocess.popen.
Below is an example of how you can do this, whilst still keeping all of the output of the program and it’s return code so that you can use it to do smart things within your python script.
Example: Running a Program with subprocess.Popen in Python
import subprocess
import logging
import os
# Configure logging
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')
logger = logging.getLogger(__name__)
def run_command(command, exit_on_fail=False):
process = subprocess.Popen(command, shell=True, stdout=subprocess.PIPE, stderr=subprocess.PIPE, text=True)
stdout, stderr = process.communicate()
return_code = process.returncode
if return_code == 0:
logger.info(f"Command succeeded: {command}")
logger.info(f"Output: {stdout}")
else:
logger.error(f"Command failed with return code {return_code}: {command}")
logger.error(f"Error Output: {stderr}")
if exit_on_fail:
# Exit if the process fails
os.exit(return_code)
return stdout, return_code
# Example usage
command = "ls -l"
output, return_code = run_command(command)
Example: logging with and error handling in bash
Logging in bash can be done using echo to print to stdout.
You can redirect output to stderr using >&2 (no spaces).
You can quit your script early with a given return code using exit.
# Log to stdout
echo "Starting analysis"
# < run a task that produces file.txt>
# Check the file was produced
if [ ! -e "file.txt" ]; then
# Log to stderr
echo "Error! <task> didn't write file.txt" >&2
# exit with return code not-zero
exit 1
fi
Bash also provides a few modes that you can enable that will help with logging and debugging.
set -xwill print each command to stdout before it is executed. Useful for tracking exactly what was executed without having to have lots ofechocommands.set -uwill treat the use of unset variables as an error. The default is that unset variables are assumed to be empty, which makes it hard to catch typos.set -ewill cause your script to exit immediately if any commands exit with non-zero status.
By having set -eux at the top of your bash script (eg workflow.sh) you’ll automatically have some basic logging and error handling in your script.
Key Points
Automation can significantly streamline repetitive tasks and reduce errors.
Creating isolated and reproducible environments is crucial for reliable research.
Effective workflow management ensures consistency and reproducibility.
Tools like
wget,venv, Make, and Nextflow are very helpful for automating and managing workflows.